Molecular Interaction Maps |
Editor(s): Allan Kuchinsky |
<<TableOfContents: execution failed [Argument "maxdepth" must be an integer value, not "[2]"] (see also the log)>>
About this document
This is an official Request for Comment (RFC) for Supporting Molecular Interaction Maps in Cytoscape.
For details on RFCs in general, check out the Wikipedia Entry: Request for Comments (RFCs)
Status
* work in progress. Consolidation of issues from use cases done and written to a preliminary Requirements section.
How to Comment
To view/add comments, click on any of 'Comment' links below. By adding your ideas to the Wiki directly, we can more easily organize everyone's ideas, and keep clear records. Be sure to include today's date and your name for each comment. Here is an example to get things started: /Comment.
Try to keep your comments as concrete and constructive as possible. For example, if you find a part of the RFC makes no sense, please say so, but don't stop there. Take the extra step and propose alternatives.
Proposal
A Molecular Interaction Map (MIM) is a diagram convention that is capable of unambiguous representation of networks containing multi-protein complexes, protein modifications, and enzymes that are substrates of other enzymes. MIMs are described in detail at http://discover.nci.nih.gov/mim/index.jsp and in a seminal article by Kohn et al at http://www.molbiolcell.org/cgi/content/abstract/E05-09-0824v1
An example MIM is below 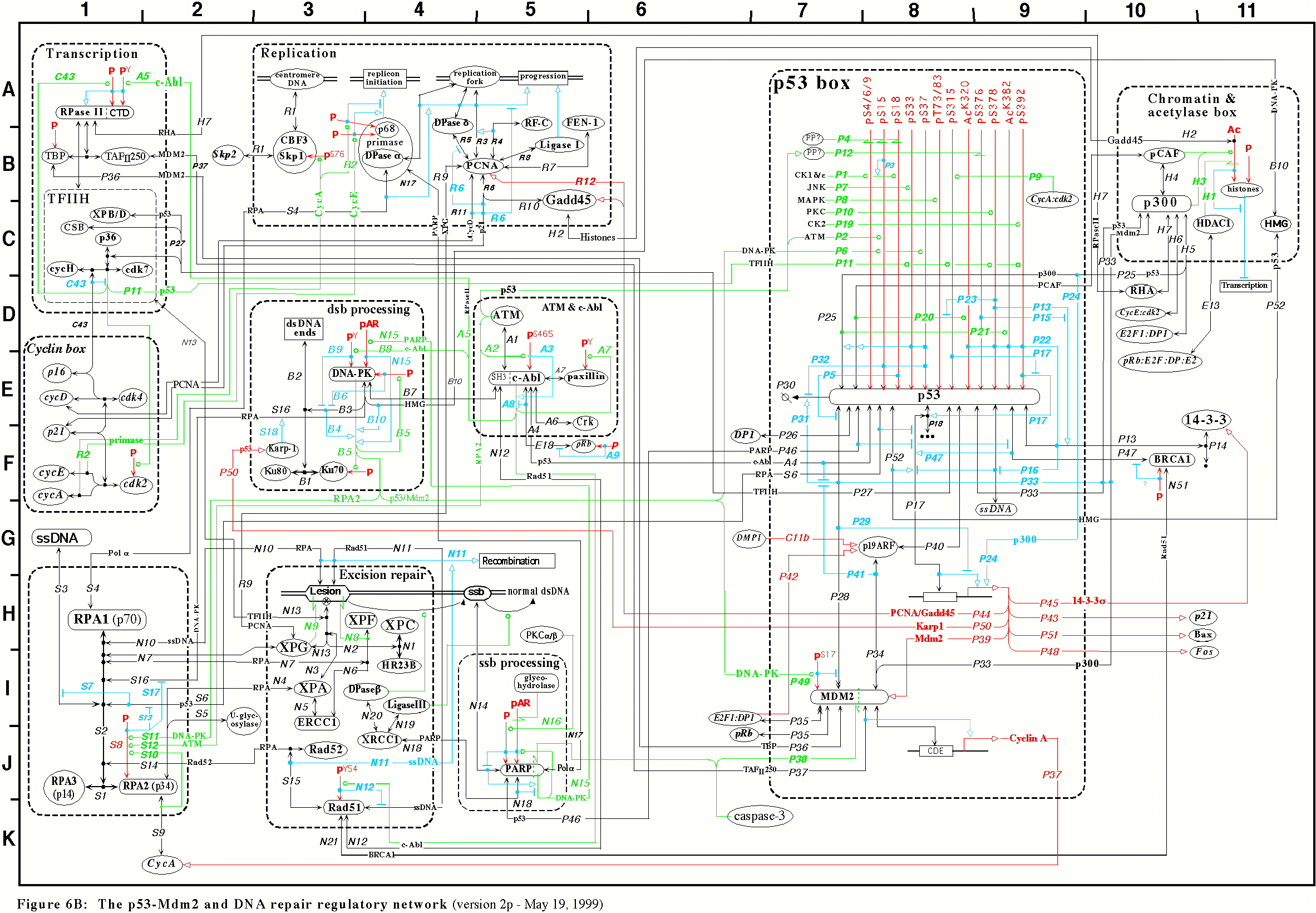
MIMs have been developed at the National Cancer Institute (specifically the Laboratory of Molecular Pharmacology). They are looking into what it would take to develop an Editor to create Molecular Interaction Maps (MIMs) in an editor to yield both a graphical view and a computer-readable format (preferrably BioPax). They are interested in seeing if such a MIM editor could be developed for Cytoscape.
MIM symbols are defined in the figure below:
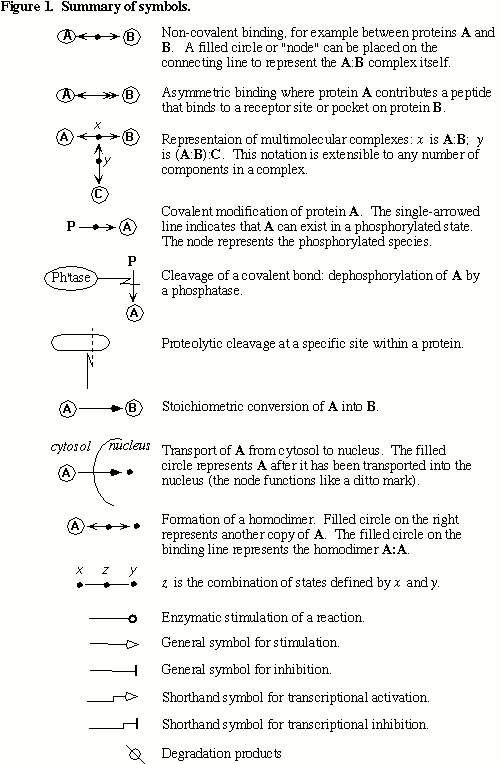
QUESTION: Is this the most recent definition of map symbols? In particular, are site-specific constructs, such as cleavage, still defined as requirements? ANSWER:There are a few changes. CovalentBinding has a new symbol. There are some requirements for site-specific constructs. There are a few additional symbols as well. This proposal will specify the mapping between MIM constructs and their proposed counterparts in Cytoscape. A prioritized set of requirements will be presented. Enhancements needed in the editor, renderer, and/or other Cytoscape components will be identified.
Kohn Notation Use Cases Editor System Use Cases
From an initial reading of the article by Kohn et al at http://www.molbiolcell.org/cgi/content/abstract/E05-09-0824v1, an initial description of potential mappings between MIM and Cytoscape constructs was derived. This is summarized in the figure below. A newer version of the description of the notation is found in Kohn et al http://mct.aacrjournals.org/cgi/rapidpdf/1535-7163.MCT-06-0640v2
The following set of requirements is drawn from a consolidation of comments on the use cases. The requirements fall into two categories: One general comment is that, for performance reasons, we may want to consider whether changes to rendering should only be implemented for high level of detail zoom factors. The major requirements are: renderer: nodes should snap to grid when being moved. InnerCanvas should display grid points, upon request from the user. Granularity of the grid and granularity of displayed grid points should be configurable by the user. When moving a Node that is attached to a segmented edge, the anchor points of the edge should remain fixed in position and the connecting sub-edge should be stretched or compressed, depending upon the direction of the movement. Implications for renderer performance: there shouldn't be too much impact for normal panning and zooming of the canvas, since nodes remain in their current coordinates and no recalculation of node coordinates is needed. However, moving a set of selected nodes within a large network could potentially affect performance of the renderer. graph layout: we should have a specialized graph layout algorithm for MiMs. The existing orthogonal layout algorithm should be a good starting point. Cytoscape desktop: the various Cytoscape modules need to know when they are in 'MiM' mode, i.e. when they need to enforce layout rules. This could be configurable by the user via an item on the Edit menu. Or it could be driven by a specialized 'MiMs' visual style. Is there a way to do this without having to add too much extra code in the editor and renderer for handling special cases. editor. It would be useful to have a library of predefined 'templates' which would handle composites such as state combination. This functionality is currently being provided in the HyperEdge editor for the use case of biochemical reactions. We should be able to build on top of that infrastructure. This does not mean that we necessarily provide functionality for the user to define their own templates. CyGroup: a number of abstractions should be represented as CyGroups. This includes dot in assymetric binding, cellular compartments, XXXX. In many situations, the GroupNode should be used to represent a MiMs grouping entity, e.g. a cellular compartment and its view. Many of these will require specialized CyGroupViewer implementations. renderer: many new CustomNodeGraphics will need to be built, e.g. to represent the boundaries of cellular compartments. HyperEdges: we ought to be able to implement snap-to-grid support in the Cytoscape editor in a way that is transparent to HyperEdges. However, this may require some support in the HyperEdgeEditor, e.g. when drawing hyperedges. readers and writers: how is information about CustomNodeGraphics output so that it can be applied again when a the MiM is read back in? Should there perhaps be a specialized BioPAX reader that knows how to handle MiMs and knows about the different CustomNodeGraphics available. Or should there be a 'View as MiM' entry on the View menu item? Perhaps we can encode the Node type as an attribute when the MiM is output, then the reader can look for this attribute on each Node that is created and do the right thing with respect to view. CyGroup: in principle, we can use CyGroups to represent these, where the group is the gene or protein and the sub-molecular entities are children of the group. This may cause complications in that the grouping exists at a different level of the network than do groups of genes and proteins. Will this cause some network analysis functions, such as shortest path, to fail? renderer: the renderer may have to render edges so that they connect to the boundary between two sub-molecular components, for example in the case of cleavage sites. We might want to introduce the context of a 'site' for a CyNode, which could be used to represent cleavage sites, splice junctions, and other sub-node entities. In the model, this could be implemented via the Group, each 'site' being a child of the group. In the view, this could be represented as a 1-pixel wide rectangle adjacent to the border of the node, or perhaps a 1-pixel square rectangle at a point on the node. This is basically a "port". readers/writers: does BioPAX represent sub-molecular entities as 'sequence features'? If so, then can a BioPAX reader be crafted to store and restore this information in the group structure. new edge types: There is a diversity of edge types that MiMs support. Implications are mostly for the renderer and editor. 'Constant' nodes: There are certain Nodes in MiMs, which represent Concepts and may appear multiple times in a network. Such 'constants' include degradation and 'mRNA'. Since Cytoscape shares nodes, we may want to represent distinct occurances of a 'constant' node as distinct entities which have the same label. Question: will this approach break many of the network analysis tools? 'Coordinate System'. This is kind of like a Legend in Cytoscape, except that it may need to update itself upon panning or zooming. Perhaps this could be implemented as a graphical annotation whose Component listens for ViewportChange events and repaints itself with the appropriate labels. handling 'first neighbor' functionality: this is required by PathBranching and PathHighlighting. Cytoscape's nearest neighbor functionality, with some minor extensions, ought to be able to handle this requirement.
Do they need site-specific connection of edges, e.g. as in proteolytic cleavage? From Mirit's presentation, it appears that metanodes would work just fine. ( see /ProteinBindingSites and /ProteinDomains) How do we represent the "null" node, as in degradation product? (see /SpeciesDegradation) How do we represent "ditto" nodes, e.g. as in translocation? (see /ShowTransport)
Biological Questions / Use Cases
General Notes
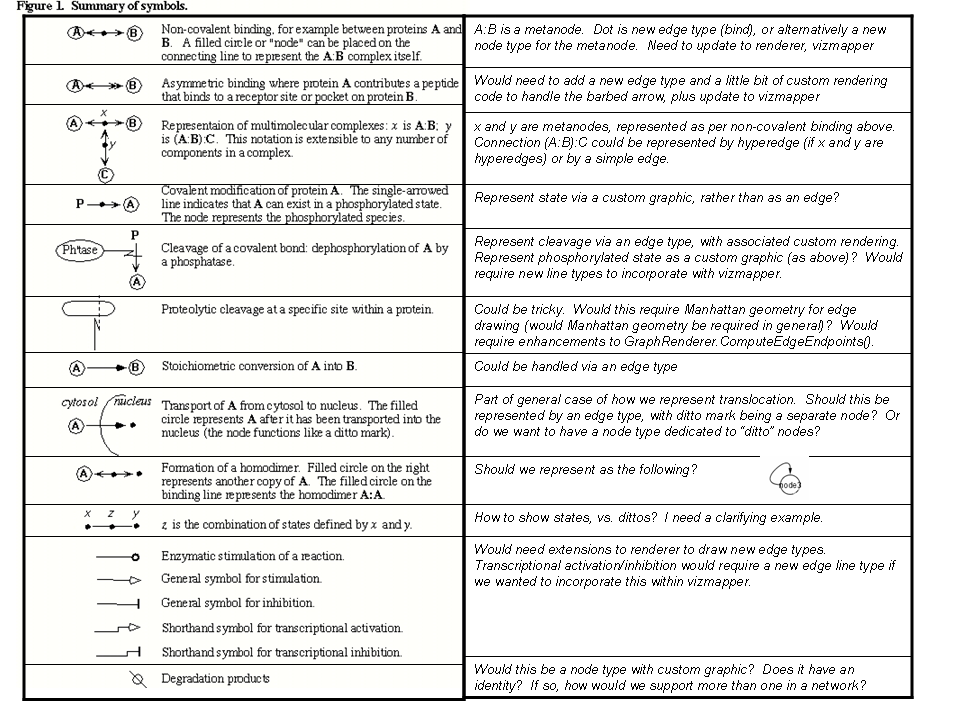
Further Notes from meeting at 2006 Cytoscape Developers Retreat
Requirements
Deferred Items
Open Issues
Backward Compatibility
Expected growth and plan for growth
References
Implementation Plan
Comments