<<TableOfContents: execution failed [Argument "maxdepth" must be an integer value, not "[2]"] (see also the log)>>
Summary
A user wants specify in trans, i.e. that a species is involved in an enzymatic or binding activity with another copy of the same species
Step-by-Step User Action
- User specifies an existing enzymatic stimulation or binding activity
User specieis that the activity is in trans
- User optionally specifies the evidence for the relationship
As an alternative to modifying an existing relationship, this use case might be viewed as creating a different relationship in the first place, e.g.
- User specifies a species to be the enzymatic stimulator
- User specifies a reaction to be stimulated involving the same species
User specifies that there is a stimulation in trans relationship
- User optionally specifies the evidence for the relationship
Visual Aides
The Kohn notation for enzymatic stimulation in trans is the following:
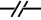
This notation can be used to modify the edges for ../NonCovalentBinding or ../EnzymaticStimulation
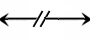

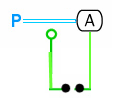
An example using this notation for ../EnzymaticStimulation follows:
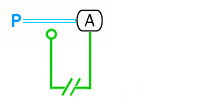
In this example, a copy of protein A, catalyzes the phosphorylation of another copy of protein A.
The following example uses intrans for a ../NonCovalentBinding relationship:
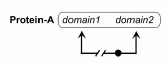
In this example, a copy of protein A, binds a domain 1 to another copy of protein A at domain 2.
Requirements for Cytoscape
Assuming that this can use the same approach that is used for ../ShowInhibition, it could be modeled as a Hyperedge containing node1: C (hyperedge attribute: enzymatic stimulation in trans), node2: A, node3: A
View: Use case for hyperedge view
Importance
This is a fairly common occurance in the models.
Variations
An earlier version of the notation uses small circles instead of the slashes. This earlier notation does not need to be supported.

Other Examples
Comments
Shared ../MimEditorUseCaseComments
This is a special case of an ../EnzymaticStimulation. It is intended to clarify that more than one copy of a particular protein is involved in the interaction.
This could be modeled in BioPAX as a Catalysis object (e.g. Catalysis object with participants = a Biochemical Reaction object and a enzyme A, Controlled = the Biochemical Reaction object (a reaction in trans as defined in "in trans reactions"cannot communicate that is specifically a covalent phosphorylation reaction, Controller=A, Control-Type = Activation, and Direction = IRREVERSIBLE-LEFT-TO-RIGHT)
AllanKuchinsky - 2007-01-20 04:29:49
model level requires new Edge type, as does view level
should there be a 'special case' of Groups in Cytoscape to represent 'sub-molecular' entities, such as domains, splice sites, etc. Is there a common enough set of semantics to justify this?
Do we ever need to represent phenomena at the level of a sequence.
MiritAladjem - 2007-02-01 09:02:47
Allan, in response to your second queston, we anticipate that people will want to use the Kohn annotation to depict interactins within molecules. We already have some people collaborating with us to develop such maps. It will be good to support this effort by allowing sub-molecular interactions. The level of a sequence might be a derivativce of sub-molecules.