Use Case Name : ../PathHighlight |
For Feature : MIMEditor |
Editors: DavidKane |
<<TableOfContents: execution failed [Argument "maxdepth" must be an integer value, not "[2]"] (see also the log)>>
Summary
A user wants to emphasize a particular portion of a map
Step-by-Step User Action
- User selects an edge in the map
- System highlights that edge, and any nodes connected to it
- Repeat steps 1 and 2 until the desired section is emphasized
Visual Aides
An illustration of what a map with a highlighted path could look like follows:
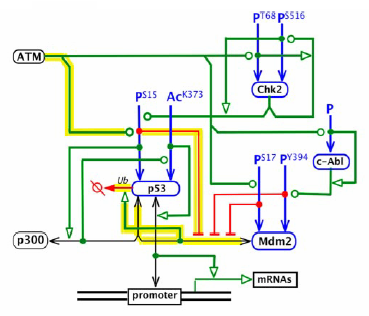
Requirements for Cytoscape
implementations (e.g., "current context menus can be used accomplish step 2 above")
TBD
Importance
This is particularly relevant when using the editor to produce images for publication.
Other Examples
Comments
Shared ../MimEditorUseCaseComments
BioPax representation TBD
AllanKuchinsky - 2007-01-23 05:23:15
At the view level, this could potentially be handled in a manner similar to hyperedges with 0-size connector nodes. But at the model level, the semantics may be inconsistent with hyperedges. A set of hyperedges with chained connector nodes implies a flow of signal from connector node to connector node. Is this consistent with the semantics of a Path with multiple branch points in MiMs?
AllanKuchinsky - 2007-05-25 10:34:16
Wait, this is simpler. We are just highlighting edges and their constituent nodes. This is similar to the PathBranching example and should be straightforward to implement.