Use Case Name : ../PathBranching |
For Feature : MIMEditor |
Editors: DavidKane |
<<TableOfContents: execution failed [Argument "maxdepth" must be an integer value, not "[2]"] (see also the log)>>
Summary
User wants to compactly layout many contigency relationships for the same species
Step-by-Step User Action
User specifies a contigency relationship (e.g. ../ProcessStimulation, ../ShowInhibition)
- User specifies a second contigency relationship of the same type with the same species origin
- User joins the two relationships with a branch
- User moves the branch point to a convenient location
Visual Aides
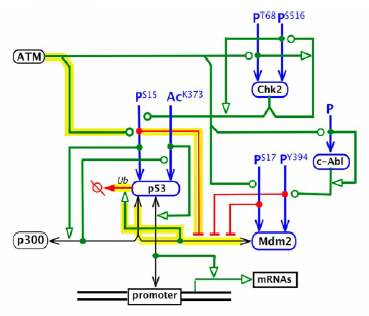
The following example has several branches, but the clearest are the several ../EnzymaticStimulation relationships from ATM. Instead of four edges going from ATM to the four species it stimulates, there is one edge that branches three times.
Requirements for Cytoscape
TBD
Importance
This important for keeping the diagrams from becoming too cluttered.
Other Examples
Comments
Shared ../MimEditorUseCaseComments
BioPax representation TBD
AllanKuchinsky - 2007-01-23 04:39:32
This is similar to the first neighbors functionality in Cytoscape (Select -> Mouse Drag Selects -> Nodes -> First Neighbors of Selected Nodes). Should be straightforward to implement.