RFC Name : Advanced Network Merge and ID Mapping API |
Editor(s): Jianjiong Gao, Keiichiro Ono, Thomas Kelder, Martijn van Iersel, Gary Bader |
Date: 2008-05-02 |
Status: open for comment |
Updates
04/02/2010: AdvancedNetworkMerge plugin was moved to core plugins for Cytoscape 2.7.0 and later.
04/09/2009: Advanced Network Merge plugin user manual.
09/15/2008: Advanced Network Merge was released. ReleaseNotes
- 07/07/2008: Attribute-Based merge function is available.
Contents
Proposal
Cytocape has implemented a plugin of Merge Networks, which allows the user to find the union, intersection and difference of networks based on node identifiers/IDs. However, nodes with different IDs may actually have the same biological meaning (e.g. the same protein/gene), especially when they come from different sources. In this project, the Merge Networks feature will be enhanced. Nodes will be matched according to ID mappings, i.e. the nodes whose IDs are mapped to each other are considered as the same node.
Use Cases
5 related use cases have been identified on Bader Lab ID Mapping page. 2 of them are closely related to this project:
Unification during dataset merging: During a merge operation e.g. of two protein-protein interaction datasets from independently created databases, it is vital to recognize that two protein objects, one from each data source, represent the same protein molecule, even if the protein objects don’t share any database accession numbers. Unification requires knowledge of record type e.g. you cannot reliably use a gene ID to unify proteins (mostly because splice variants exist).
Identifier translation: Some analysis methods require specific translations from one set of identifiers to another. For instance, our 'activity centers' analysis requires translation from protein or gene identifiers in a pathway database to Affymetrix probe set identifiers or other gene expression array platform identifiers.
Workflows
Main procedure
Given a set of source networks, each of which has a set of attributes, Network Merge procedure should have the following three steps:
- Node matching: select attribute(s) of each source network to identify the nodes in the network. Two nodes in two networks match when the value of their selected attribute(s) match (i.e. map to each other if using ID mapping, or the same if node). Note: the main purpose of this step is to matching nodes among source networks. Using ID mapping is just for matching nodes in this step.
- Attribute merging: merge the attribute of source networks into attributes in the resulting network. The user can define the attributes in the resulting network: the names of the attributes and which attribute in which source network it comes from. The user can define as many or as few attributes as he/she likes. If the values of attributes of the matched nodes are the same, then merge without problem; otherwise, conflicts occurs. Note: ID mapping can be also used in this step to match values of attributes of the source network.
- Conflicts handling: let the user decide some rule based on priorities of networks, priorities of ID types, etc.. Or let the user assign which attribute value should be used for each node. Note: using IDs of a destination id type to assign the IDs in the resulting network is actually one of the strategies to solve the conflicts of IDs among different ID types.
ID mapping procedure
- Selecting which ID mapping source to use. Below are the possible options from which the user can choose.
- Provide custom mapping with a local file
- Get ID mappings from a web service
- Get ID mappings from a relational database (e.g. embeded Derby database)
- Selecting which ID types are used in the networks from a list of ID types.
- The list of ID types may be different for different database or web service. For example, if the user chooses GenMAPP gene database, he/she can select a few out of the 19 supported ID types.
Or according to GaryBader's suggestion, we should limit the types of IDs supported in order to minimize the chances for errors. On that purpose, we should only provide the most common used ID types (e.g. Entrez Gene, RefSeq, UniProt and some others) in the list of ID types, no matter what web service/database the user has chosen.
- If the user is not sure about what ID types are used in the networks, he/she can choose all.
After retrieving the data, report to the user how many IDs are found, how many are not and how many are ambiguous (the same ID existing in different type--should be rare). For the ambiguous ones, ask the user to decide. (This is similar to what DAVID has done.)
A simple case study
There are two source networks Net1 and Net2. Net1 has two attributes ID and Name; Net2 also has two attributes ID and Alias. There are 3 nodes in Net1 and 2 nodes in Net2. ID in Net1 belongs to ID type 1 and ID in Net2 belongs ID type 2.
Networks
Net1
Net2
Attributes
ID
Name
ID
Alias
ID type
type 1
type 2
Node
1
Gene1
01
Gene1
Node
2
Gene2
02
Gene02
Node
3
Gene3
In step 1 of the main procedure, the user selected ID in Net1 and ID in Net2 as a pair of attributes to match nodes. By some ID mapping service shown as below, ID of node 1 in Net1 and ID of node 01 in Net2 are both mapped to ID 001 of ID type 3; ID of node 2 in Net1 and ID of node 02 in Net2 are both mapped to ID 002 of ID type 3; ID of node 3 in Net1 cannot be mapped to type3.
ID
Source type
ID
Destination type
1
type 1
001
type 3
2
type 1
002
type 3
3
type 1
N/A
type 3
01
type 2
001
type 3
02
type 2
002
type 3
In step 2, the user defined the attributes in the resulting network in the table below. Now the task is to merge the attributes of the matching nodes: node 1 in Net1 to node 01 in Net2 and node 2 to node 02. If merging according the same value of attributes, only node 3 in Net1 successfully merged into the resulting network without conflicts (only exist in Net1, no chance:). Attribute Name in Node 1 and Alias in Node 01 are merged in to Name in Net_res without conflict, but ID in Net1 and ID in Net2 have conflict. For Node 2 and Node 02, both attributes have conflicts.
Net1
Net2
Net_res
ID (type 1)
ID (type 2)
ID (type 3)
ID(type1)
ID1 (type 1)
ID (type 2)
ID2 (type 2)
Name
Alias
Name
In step 3, the user chose to use ID mapping and ID type 3 as ID type of the resulting network. So node 1 and 01 are merged to a node with ID 001, and node 2 and 02 to 002. For node 3, it cannot be mapped to type 3, so the original ID is taken as ID of the resulting node. Also node 3 in the resulting network has no value for attribute ID2. For attribute Name in the resulting network, the user chose the Name in Net1 if conflicts occur. Finally, a resulting (union) network could be a network as below.
Networks
Net_res
Attributes
ID
ID1
ID2
Name
Node
001
1
01
Gene1
Node
002
2
02
Gene2
Node
3
3
Gene3
Assumptions
Because of internal architecture of Cytoscape, we assume the following for merge operation:
Attribute called Canonical Name always exists.
If not, it should be created before merge operation. This can be done by simply copy all Node IDs into attribute Canonical Name.
Canonical Name is always identical to Node ID
If user wants to merge based on Node ID, use Canonical Name instead
Merged network has nodes with newly-generated Node ID
- Example 1 - the following information represents a same node in two different networks:
Network 1
Network 2
Node ID
4000
HGNC:6636
Canonical Name
4000
HGNC:6636
Common Name
LMNA
(Not available)
Official Symbol
(Not Available)
LMNA
User wants to merge them based on Common Name and Official Symbol. After merge operation, the new node looks like the following:
Merged Network
Node ID
4000-HGNC:6636
Canonical Name
4000-HGNC:6636
Entrez Gene ID
4000
HGNC ID
HGNC:6636
Name
LMNA
Common Name
LMNA
Official Symbol
LMNA
Name attribute is the newly created attribute by matching Official Symbol and Common Name. Name of the new attribute is user editable in the GUI and this operation is optional. User need to name enter attribute names Entrez Gene ID and HGNC ID. These are created from Canonical Name in the original network. By default, they are set to Canonical Name: TITLE_OF_NETWORK_1 and Canonical Name: TITLE_OF_NETWORK_2
Creating new node is not always necessary if they share the same ID in the original networks. For example, if we merge the following two nodes in two different networks based on Canonical Name,
the result is the following:Network 1
Network 2
Node ID
ENSG00000160789
ENSG00000160789
Canonical Name
ENSG00000160789
ENSG00000160789
HPRD ID
HPRD:01035
(Not available)
Official Symbol
(Not Available)
LMNA
Merged Network
Node ID
ENSG00000160789
Canonical Name
ENSG00000160789
HPRD ID
HPRD:01035
Official Symbol
LMNA
- In this case, merge operation does not create new node.
- Example 1 - the following information represents a same node in two different networks:
Examples
Implementation Plan
General Architecture
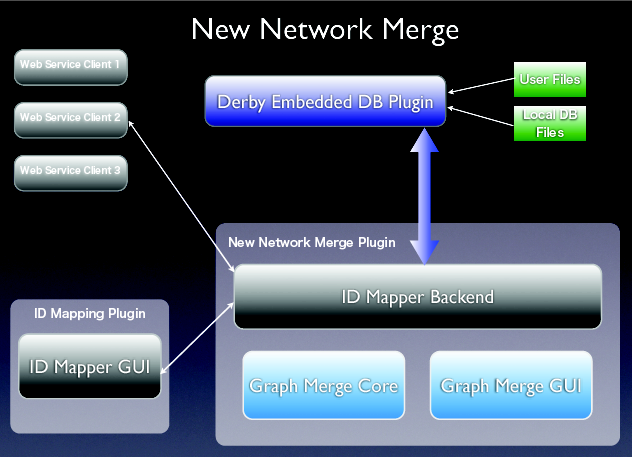
- This project will be devided into 3 parts.
- ID Mapping Module
Implements function to map one ID set (UniProt, NCBI Gene ID, etc.) to the other.
- This function should be accessible from other modules (plugin). Public API will be published for other plugin developers.
- Database Module
- Wrap Derby with a genetic interface for Cytoscape.
- Storing mappings locally.
- Data from remote sources (web services) will be cached here.
- Network Merge Module
- Actually merging multiple networks.
- Dealing with attribute conflicts.
- This includes new GUI.
- ID Mapping Module
- The following module is optional. Will be implemented if we have time.
- ID mapping validator
Code Base
- Based on Cytoscape 2.6.x branch since 3.0 will not be available until next year.
- However, we should try to minimize the amount of work for porting to 3.0 series. This should be done by interface-based design, i.e., design clean API of rthe outside of the world and should be useable by other plugin developers.
- Interoperability between modules (plugins) should be considered.
GUI Design
Some Requirments Based on Other Developer's Feedback
- Type-Guessing - Use regular expression or number of exact match to guess ID type. With this function, users do not have to select ID type (in most cases).
- Matching nodes based on multiple attributes
API Design
API for ID mapping
Basically, the interface takes a list of orignal IDs, original ID types and destination ID type as parameter and return a list of destination IDs.
1 public interface IDMapper {
2 /**
3 * Map a list of IDs of one type to a list of IDs of another type
4 * @param idSrc The list of source IDs, i.e., the list of IDs to be mapped
5 * @param typeSrc The type (e.g. UniProt ID) of the source IDs
6 * @param typeDst The destination ID type
7 * @return a list of distination IDs
8 */
9 public List<String> mapID(List<String> idSrc, String typeSrc, String typeDst);
10 }
11 public interface IDMapperDB extents IDMapper {...}
12 public class IDMapperDerby implements IDMapperDB {...}
13 public class IDMapperWebservice implements IDMapper {...}
14 public class IDMapperFile implements IDMapper {...}
15
- Question:
- Interface VS abstract Class?
- Alt. design:
- Instead of String, we may need to use enum to define supported id types.
- ID Types will be defined as Strings to make the list expandable.
- Basic interface for ID Mapping API.
1 public interface IDMapper {
2 // Supports one-to-one mapping and one-to-many mapping.
3 public Map<String, Set<String>> mapID(Set<String> ids, String srcType, String tgtType);
4
5 // Check whether an ID exists in a specific type.
6 public boolean idExistsInSrcIDType(String srcID, String srcType);
7
8 // returns supported source ID types
9 public Set<String> getSupportedSrcIDType();
10
11 // returns supported target ID types
12 public Set<String> getSupportedTgtIDType();
13 }
14
- The basic Mapper interface will be extended to add some more methods for each datasource types. Supported datasources are:
- Mapping file
- Relational database
- Web service
1 public interface IDMapperFile extends IDMapper {}
2 public interface IDMapperRDB extends IDMapper {}
3 public interface IDMapperWebService extends IDMapper {}
4
- Implementations for each data source
1 public class IDMapperText extends IDMapperFile {
2 // Delimited text file mapper implementation
3 }
4 public class IDMapperExcel extends IDMapperFile {
5 // Excel file mapper implementation
6 }
7 public class IDMapperDerby extends IDMapperRDB {
8 // Apache Derby specific mapper implementation
9 }
10 public class IDMapperMySQL extends IDMapperRDB {
11 // MySQL specific mapper implementation
12 }
13 public class IDMapperUniprotWS extends IDMapperWebService {
14 // Uniprot web service specific mapper implementation
15 }
16 .
17 .
18 .
19
Comments on API
KeiichiroOno (2008-05-02): It (API for ID mapping) should be as simple as possible; just takes original ID (or ID set), original ID type (entrez gene id, uniprot id, etc.), and destination ID type.
GaryBader (2008-05-03): I think the default query should be as Kei suggested, but some users will want to merge in specific ways (e.g. unification in one species vs. cross-species), so having an optional 'id mapping type' controlled vocabulary could be useful. I would suggest creating some test cases of actual different types of ID mappings that users typically do - these could be used to test the generality of the API.
PietMolenaar (2008-05-05): Would it be possible to introduce an additional interface IDMapperDb of which IDMapperDerby is a sample implementation, in order to facilitate generalized mapping to relational databases
KeiichiroOno (2008-07-07): Interface design updated. Instead of hidden abstract classes, extended interfaces will be public and accessible from plugin writers. (Still, actual implementations are hidden.) Need to define required methods for each data source.
File format
- Free format text table file or MS Excel file (ref: import network from table)
- Each column for one ID type
- Each row except the first one represents IDs of different types mapping to each other
- First row contains ID types
- Multiple IDs are allowed to be contained in one cell (One to many mapping, or IDs of the same type maps to each other). Use special character (e.g., ';', '/', etc, or user defined) to separate IDs.
- Relational database tables to store ID mapping
- Two solutions:
1. Three tables similar as in PathVisio:
DataNode (ID, Type): each tuple contains the ID of a node and its ID type.
- Link (ID1,ID2): each type contains two IDs mapping to each other.
- Info (Version): containing the version of data.
- 2. Two tables:
- Data(VID,ID,Type): each tuple contains a virtual ID, the ID of a node and its ID type. IDs mapped to each other have the same virtual ID.
- Info (Version)
- Data(VID,ID,Type): each tuple contains a virtual ID, the ID of a node and its ID type. IDs mapped to each other have the same virtual ID.
- Question:
- Which solution to use?
- How to store cached data? Separate tables or additional fields?
- Two solutions:
References/Resources
Related work
5 related use cases identified on Bader Lab ID Mapping page
PathVisio synonym databases store the mappings from Ensembl using Derby database ( package & src--GDB classes in the org.pathvisio.data)
Web services and Relational DB
Tools
ScriptingPlugins can be utilized to test web services before writing actual Java code. Ruby plugin has SOAP utilities and BioRuby inside.
NetBeans IDE: nice tool for mocking up GUI. It has a visual GUI editor for Swing. Also, it has tools to develop web service clients easily from GUI
Project Management
Project Timeline
Overall schedule for Google Summer of Code is available here.
Provide a timeline for implementation. Insert a graphic if you can. Try this free online tool for making project timelines -> Help-u-Plan (create a new chart; modify; right-click to save gif; then attach to this page)
Tasks and Milestones
This project will use incremental approach.
Outline the major milestones and tasks involved in implementation.
Tentative Schedule
Milestones
Define ID Mapping API
Design overall system structure and moch UI
Implement existing attribute-based network merge - First version available. Advanced Network Merge plugin jar file
Finish File-Based ID Mapping
RDB-based ID Mapping
Web Service Based ID Mapping
Integration and Testing
Documentation and Public Release
- Optional Tasks
- XML file readers for ID import
- Use multiple ID data sources for mapping at once
Project Dependencies
Another GSoC project taken by ArmanAksoy is also Advanced Network Merge. His approach is different: he proposed to merge nodes according to similarity score rather than ID mappings. However, for the GUI part and the part of deciding attribute conflicts, these two projects can share a common mechanism.
Related RFCs
RFC 29: Web services API for ID mapping/translator service
RFC 39: Cytoscape Data Integration
RFC 45: Web Services Client Manager and Unified Network/Attribute Import Mechanism
Issues/Discussions/Further investigations
- How to design the public interface for ID mapping function? How should Derby DB, Web service clients and ID mapper interact with each other?
- Better to keep the ID mapping interface simple: it just takes original ID, original ID type, destination ID type as parameters and returns destination ID.
- Implementation of the ID mapping interface should take the ID mapping sources (Derby DB, custom file and web service) into consideration
- If the user chooses Derby DB to get ID mapping, just search local Derby DB and return the result.
- If the user chooses custom file or web service,
- Try to search the data in Derby first
- For IDs not found in Derby, try custom file or web service
- Cache the ID mappings retrieved from web service into Derby (merge data in Derby may be needed)
- Should we implement a number of validations to detect mistakes in the ID mappings? How? -- Need further investigation.
GaryBader: It would be good to have a validation and cleaning system that implements a number of validation rules to detect mistakes in the input ID mapping files. Bad ID mappings downloaded from external sources can cause errors in the network merge, which will degrade the data. For instance, if you download uniprot and extract all of the ID mappings, you will find that some of the supposedly equivalent IDs actually point to proteins from different species. So organism, molecule type (rna, dna, protein, gene), etc. should be part of the system and associated with each ID to enable better validation.
GaryBader: What I'm thinking here is a module (separate from Cytoscape) that validates and cleans ID mappings gathered from different relatively trusted sources e.g. Ensembl, uniprot, entrez gene, refseq. We've found ID mapping errors in these sources, so it would be useful to create a cleaned set for our users. This could be a separate module to simplify the main API i.e. the main API could assume that the IDs are clean and will simply use them as is. This would also simplify the input formats (text file or database), since they would just need ID1:DB1, ID2:DB2, ID mapping type. If large file size is an issue, these could be compressed in interesting ways given there is a lot of repetition in IDs i.e. all human ensembl IDs start with ENSP000.. and you can convert the strings to integers (but this optimization is getting ahead of ourselves and should be the last thing considered, only if necessary)
- For ID mappings, should we limit the types of IDs supported?
GaryBader: The user should be allowed to use whatever IDs they want, however we may only want to provide a limited 'suggested' or 'recommended' subset in any files or databases we provide to encourage users to use more consistent IDs in their work and reduce errors and our maintenance costs.
- Should it be required that one network can only contain one type of IDs?
ThomasKelder: Maybe there are also different options here, depending on the network type. Sometimes the ID type is fixed for an attribute (e.g. an attribute called 'uniprot' typically only contains uniprot IDs), but sometimes the type is defined in another attribute (e.g. an attribute 'ID' and 'system') or concatenated to the ID (e.g. 'Uniprot:U1234'). It will be hard to cover all these options, maybe some investigation is needed on what situations occur most.
KeiichiroOno: Ideally, each network contains only one ID set. However, in some cases it is impossible since one ID set does not contain all of the macro/small molecules (see this example from IntAct. Node IDs are UniProt unified acc. number, but it is not available for small molecules). Just like IntAct, probably it is OK to use DBName:ID style ID if the object does not exists in the target ID set. In that case, the DBName:ID should be taken from the primary source of the database, i.e., if the object only exists in KEGG and not in UniProt, it should be KEGG:xxxxx
- How to maintain a Derby database on the server and client? is it affordable?
- For very large ID lists, it may take too much time to get ID mappings from web service, a local database may save a lot of time. The issue is that effort may be needed to maintain/update such a database and the Cytoscape need to update the local database (either manually or automatically) from time to time. Is it affordable?
An possible way may be like this: we maintain an ID mapping database/file on the server, update it every a few days from online web services/databases (e.g. NCBI, UniProt). The Cytoscape user can manually download/update it. when the user chooses to get ID mappings from web services/online databases, search the local database first, and then for the IDs that are not contained in the local database, try to get them from online web services/databases, if successful, save these ID mappings in the local database. The advantage of the method is that only the used IDs will be updated in the local database, rather then the whole database (though, the user can manually update it). This is desirable, because most likely one user will use a subset of the IDs. It's not necessary to update the whole database.
One question: Are the ID mappings changeable? Or does updating ID mappings mean only adding new mappings or possibly change the old mappings? If the ID mappings are changeable, it's better to add a time attribute for each ID mapping in the database, so we can update the old (e.g. >3 months) ID mappings in the local database (only update the used the IDs when searching).
- Even if we do not maintain an ID mapping database on the server, it is still good to maintain a local database/file, which contains the ID mappings the user has used, so that the user do not need get them from web service every time. It may be especially desirable, when a tool for mapping IDs is implemented.
Comments
KeiichiroOno (2008-04-22):
- Separate Derby as embedded database plugin and make it accessible from other part of Cytoscape (for flexibility and reuse).
- Caching retreved ID sets from web services and from the next query, try local DB first for performance.
- Design some GUI mockups and checks feasibility.
For local database, we may need to start 3 big tables with the following primary key: NCBI Gene ID, Ensembl Gene ID, and UniProt unified accession number. These three covers a lot of objects in biological databases.
Add comment here…
How to Comment
Edit the page and add your comments under the provided header. By adding your ideas to the Wiki directly, we can more easily organize everyone's ideas, and keep clear records. Be sure to include today's date and your name for each comment. Try to keep your comments as concrete and constructive as possible. For example, if you find a part of the RFC makes no sense, please say so, but don't stop there. Take the extra step and propose alternatives.