This feature is deprecated
Please use cyREST instead.
Scripting Engines for Cytoscape
Status
Dec. 7, 2009: All scripting engines are updated to the latest version. (KeiichiroOno)
Contents
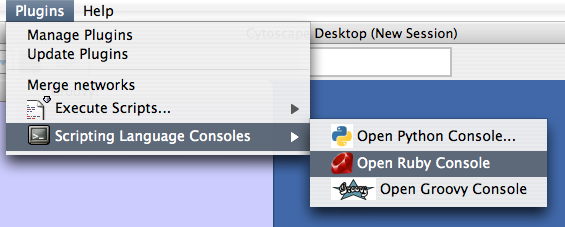
This document is for experimental implementation of Cytoscape scripting framework.
To use scripting engines, you need the following:
- Cytoscape 2.6.3 or later.
ScriptEngineManager plugin 0.08 or later
Please restart Cytoscape after you install Scripting Engines from Plugin Manager
You can run scripts from Plugins → Execute Scripts... → (engine name).
These plugins are still experimental, but if you have a small problems (like import hundreds of sif files and do some simple calculation) and if you need a quick solution, this feature is useful.
Groovy (New!)
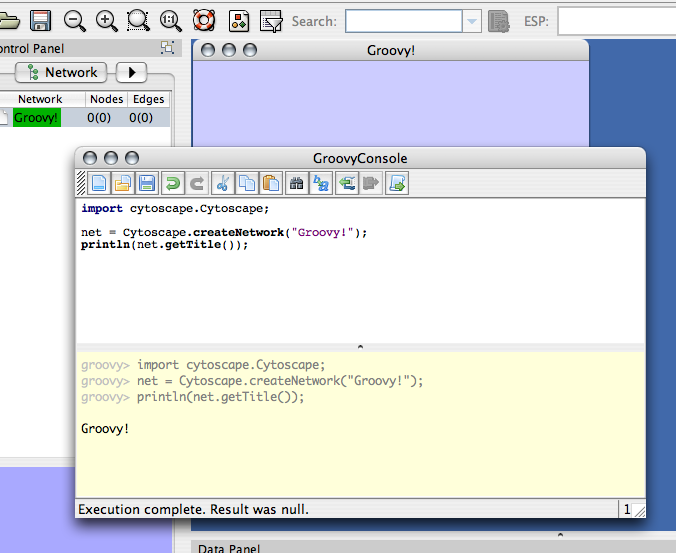
This plugin is an execution environment and interactive console for Groovy scripting language. Since this language is designed for Java programmers, the syntax is almost same as Java. If you are a Java programmer and want to use dynamic languages with low learning cost, Groovy is a good choice.
Sample Scripts
Python (New!)
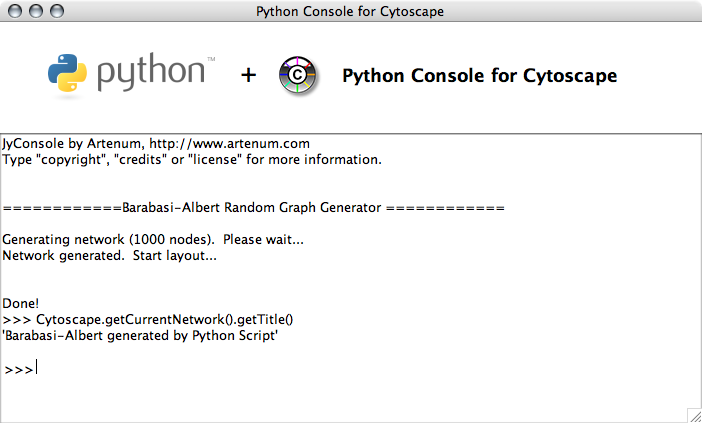
This engine uses JyConsole to run Python scripts on Cytoscape. You can use the console as the standard output of your script files.
Sample Scripts
BA Random Graph
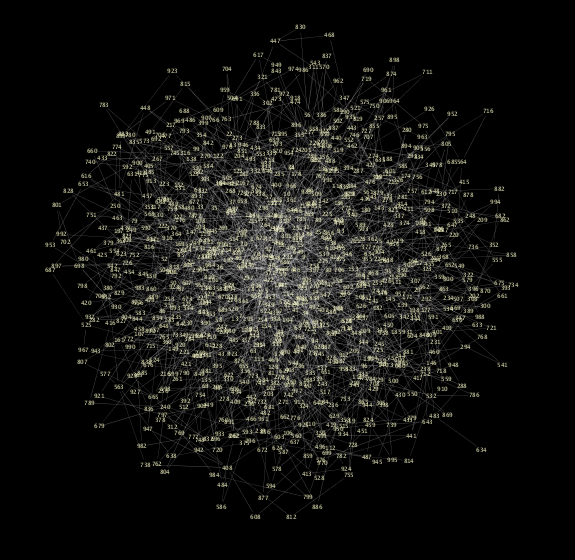
Random graph generator ported from Ruby samples below.
Ruby
Important Note
From RubyScriptingEngine version 0.21, you need to install JRuby 1.4.0 by yourself.
Go to JRuby web site and download JRuby binary: http://jruby.org/download
- Windows user - Just run Windows Installer.
Mac/UNIX users - Extract the archive and set the $JRUBY_HOME environment variable.
- To test the JRuby, open the terminal.
Type the following command and make sure $JRUBY_HOME is set.
kono$ echo $JRUBY_HOME /Users/kono/Documents/jruby-1.4.0
Set your PATH to $JRUBY_HOME/bin
Install latest version of BioRuby
gem install bio
- If you want to use other gems with Cytoscape, please install them, too.
This engine is based on JRuby technology, which is a ruby implementation on Java virtual machine. You can access Cytoscape's objects directly from your ruby script. Since ruby is an object-oriented programming language, it is relatively easy to mix objects from Cytoscape (java) and ruby.
Sample Scripts
To run the following scripts, please install RubyScriptingEngine plugin 0.21 or later.
Hello World
require 'java'
include_class 'javax.swing.JOptionPane'
JOptionPane.showMessageDialog(
nil, "Hello Cytoscape!", "JRuby on Cytoscape",
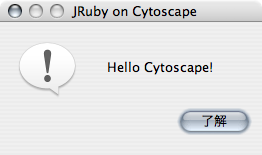
JOptionPane::INFORMATION_MESSAGE)This code segment displays a dialog like the following:
The point is, if you want to access Java classes, you need to add:
require 'java'
Load and Layout Networks
This scripts load all sif network files in sampleData directory. Also, force-directed layout will be applied for each networks with view.
require 'java'
include_class 'cytoscape.Cytoscape'
include_class 'cytoscape.layout.CyLayouts'
include_class 'cytoscape.layout.LayoutAlgorithm'
include_class 'cytoscape.CytoscapeInit'
props = CytoscapeInit.getProperties
props.setProperty("layout.default", "force-directed")
Dir::glob("sampleData/*.sif").each {|f|
puts "Loading: " + f
Cytoscape.createNetworkFromFile f
Cytoscape.getCurrentNetworkView.redrawGraph(false, true)
}
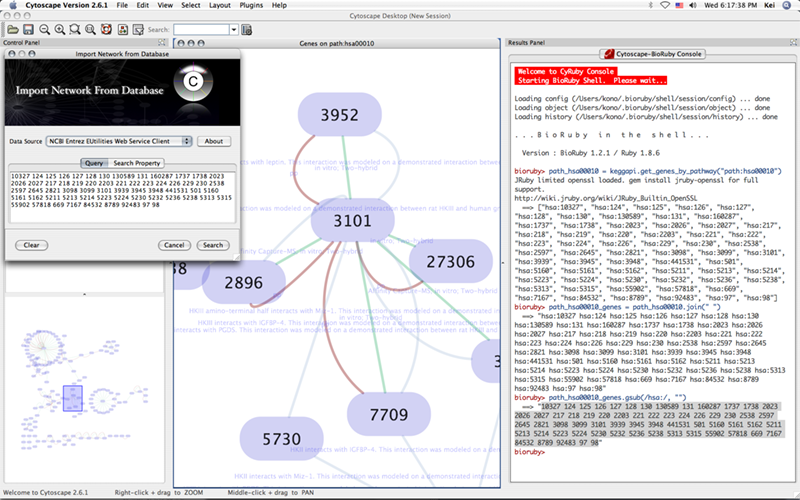
Access Remote Database by BioRuby
Now let's try some more realistic problem. The following script converts NCBI Gene ID into KEGG ID, and then import KEGG Pathway ids for each node if it exists on known KEGG pathways. BioRuby is used to access KEGG API.
This may take a long time to finish, so try small network first.
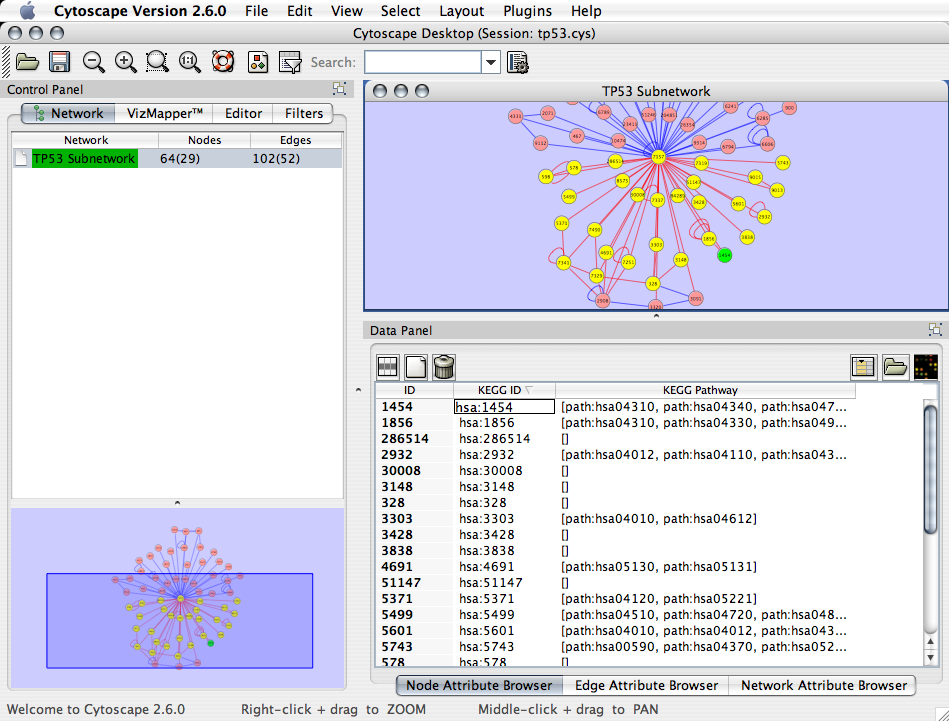
Generate Scale-Free Random Network by Barabasi-Albert Model
Scripting is useful when you want a prototype implementation of your algorithms. The following is a ruby script to generate a random graph based on Barabási-Albert model.
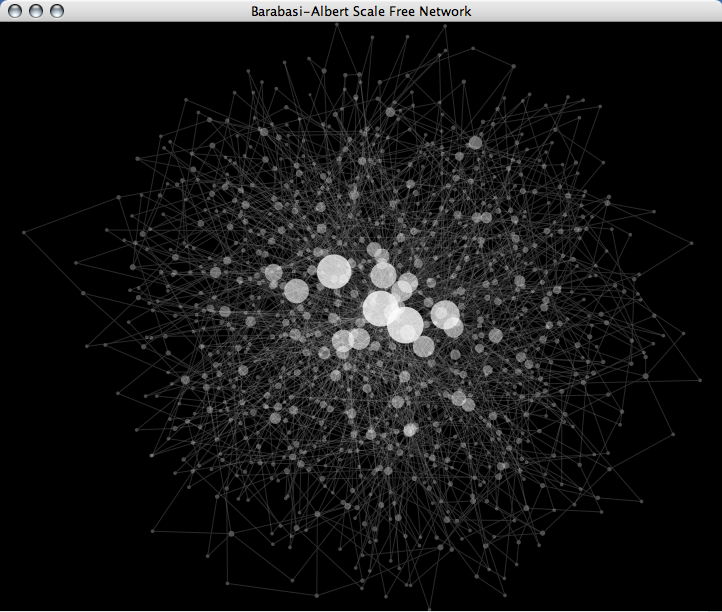
(In this Visual Style, node size is mapped to degree of node. You can see some hubs in the network, which is a character of scale-free networks.)
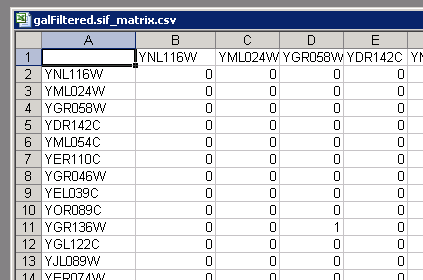
Binary Connection Matrix
The following script exports current network as binary matrix (CSV)
require 'java'
include_class 'cytoscape.Cytoscape'
include_class 'cytoscape.CyNetwork'
include_class 'cytoscape.CyNode'
net = Cytoscape.getCurrentNetwork
source = net.nodesList
line = ','
source.each do |column|
line << column.getIdentifier << ','
end
fileName = net.getTitle + "_matrix.csv"
File.open(fileName, "w") {|file|
file.puts line
source.each do |s|
line = s.getIdentifier + ','
targets = net.nodesList
targets.each do |t|
if net.edgeExists(s, t)
line << '1,'
else
line << '0,'
end
end
file.puts line
end
}
Interactive Console
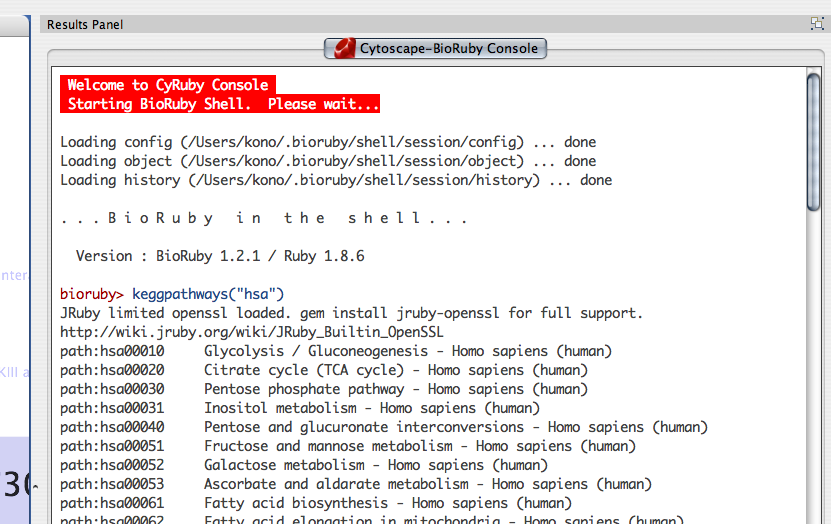
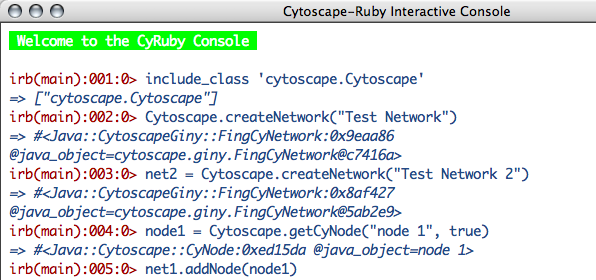
From version 0.03, you can control Cytoscape from interactive command line. You can open the console from Plugins → Open Ruby Console.... You can use TAB key for coding assistance. This feature is still experimental, so please let me know if you notice problems.
New ! --- BioRuby Shell is integrated to the console from v0.10.
P.S. I'm not a ruby programmer, so please let me know if you have any idea to improve the scripts above (Kei)
JavaScript
This plugin is based on Rhino JavaScript engine. You can execute JavaScripts on Cytoscape.
Sample Scripts
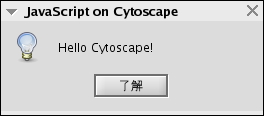
Hello World
importPackage( Packages.javax.swing );
JOptionPane.showMessageDialog(
null, "Hello Cytoscape!", "JavaScript on Cytoscape",
Packages.javax.swing.JOptionPane.INFORMATION_MESSAGE
);
Accessing Cytoscape Objects
You can access Cytoscape object from simple scripts:
importPackage( Packages.javax.swing );
importPackage( Packages.java.lang );
importPackage( Packages.cytoscape );
dialog = new JDialog();
dialog.setAlwaysOnTop(true);
dialog.setTitle("JavaScript Output");
editor = new JEditorPane();
editor.setContentType("text/html");
builder = new StringBuilder();
builder.append("<html><body>");
builder.append("<h4><font color=\"#ff0000\"> Status of Current Network: " + Cytoscape.getCurrentNetwork().getTitle() + "</font></h4>");
builder.append("<ul><li>" + "Number of Nodes = " + Cytoscape.getCurrentNetwork().nodesList().size() + "</li>");
builder.append("<li>" + "Number of Edges = " + Cytoscape.getCurrentNetwork().edgesList().size());
builder.append("</li></ul>");
builder.append("</body></html>");
editor.setText(builder.toString());
dialog.add(editor);
dialog.pack();
dialog.setSize(300, 150);
dialog.setLocationRelativeTo(Cytoscape.getDesktop());
dialog.setVisible(true);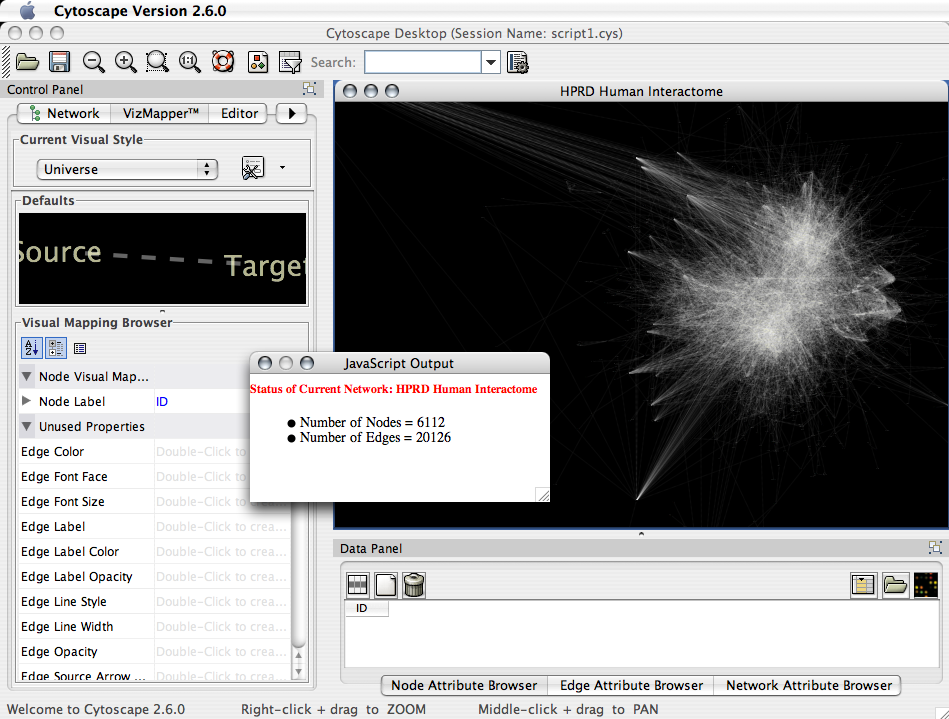
Generate Graph
The following example generates a complete graph and then apply force-directed layout. If you want to create random graphs based on your algorithms, use the following code segment as a template.
// Generate a complete graph
importPackage( Packages.javax.swing );
importPackage( Packages.cytoscape );
importPackage( Packages.cytoscape.layout );
newNetwork = Cytoscape.createNetwork("Complete Graph 1");
var nodes = new Array();
for (i=0; i<10; i++) {
nodeName = "Node " + i;
nodes.push(newNetwork.addNode(Cytoscape.getCyNode(nodeName, true)));
}
for (i=0; i<10; i++) {
for (j=0; j<10; j++) {
if(i != j) {
edge = Cytoscape.getCyEdge(nodes[i], nodes[j], "interaction", "pp", true);
newNetwork.addEdge(edge);
}
}
}
Cytoscape.getCurrentNetworkView().redrawGraph(false, true);
CyLayouts.getLayout("force-directed").doLayout();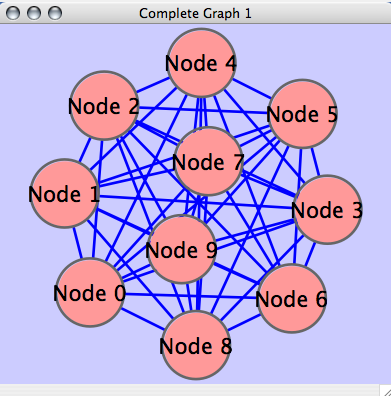
Use Google Social Graph API (Web Service)
Scripting is useful when you want a quick & dirty solution. The following is a script to visualize relationship between a person in Twitter and Followers using Google Social Graph API.
// Visualize Relationship in Twitter by using Google Social Graph API
importPackage(java.io);
importPackage(java.net);
importPackage( Packages.cytoscape.layout );
importPackage( Packages.cytoscape );
// Change this to your id
var myURL = "http://twitter.com/c_z";
var newNetwork = Cytoscape.createNetwork("Twitter Graph");
var me = newNetwork.addNode(Cytoscape.getCyNode(myURL, true));
var nodes = new Array();
var url = new URL('http://socialgraph.apis.google.com/lookup?q=' + myURL + '&edo=1');
var stream = new BufferedReader(new InputStreamReader(url.openStream()));
var line, json = '';
while(line = stream.readLine())
json += line;
stream.close();
relations = eval("(" + json + ")");
var people = relations.nodes[myURL].nodes_referenced;
for(key in people) {
if(people[key]['types'] != 'me')
nodes.push(newNetwork.addNode(Cytoscape.getCyNode(key, true)));
}
for(i=0; i<nodes.length; i++) {
edge = Cytoscape.getCyEdge(me, nodes[i], "interaction", "contact", true);
if(edge != null)
newNetwork.addEdge(edge);
}
Cytoscape.getCurrentNetworkView().redrawGraph(false, true);
CyLayouts.getLayout("force-directed").doLayout();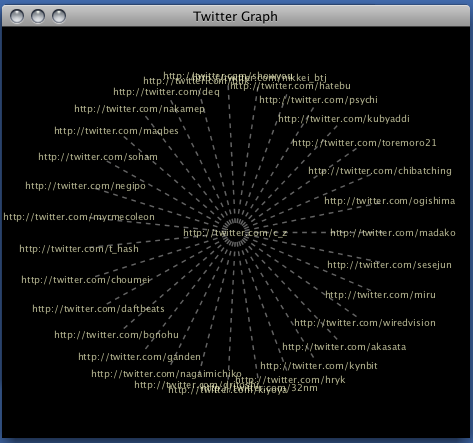
(Universe Visual Style applied.)
Of course, you can modify the script to do a bit more meaningful. The following is a script to search up to friends-of-friend (search depth 1).
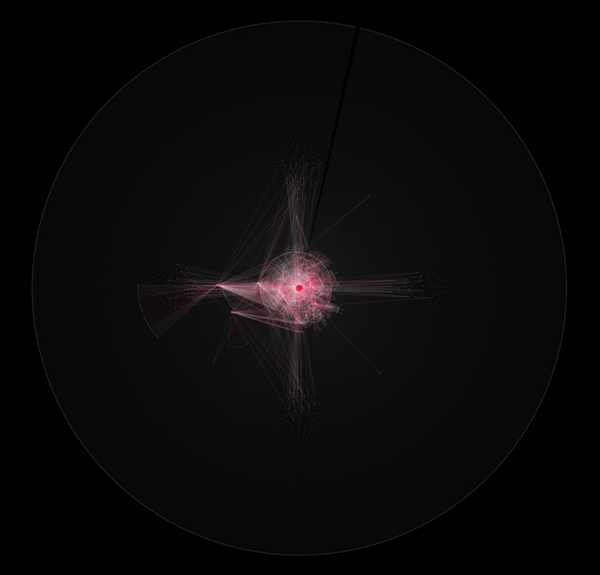
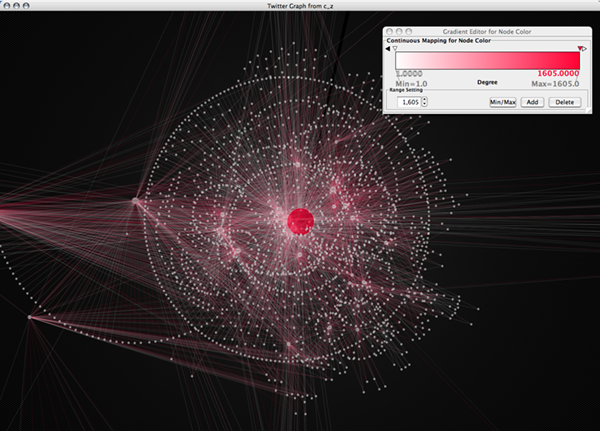
Like this example, you can write small web spiders quickly with scripting language feature in Cytoscape.