Web Service Client Reference Implementations
The following is the sample implementation of Web Service Clients for Cytoscape. Example workflow created by these plugins are available at WebServiceWorkflow page.
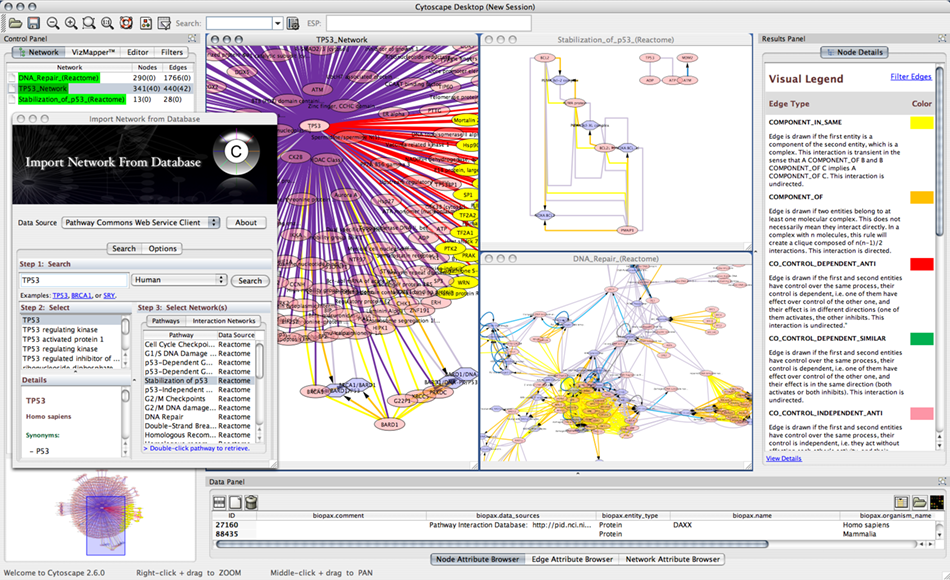
Pathway Commons Client Plugin
General Information
Database Web Site |
|
WSDL |
n/a |
Data Type |
Protein-Protein Interactions and Biological Pathways |
Pathway Commons Client is a program to access Pathway Commons database through its Web Service API. Pathway Commons is a collection of publicly available pathways from multiple organisms. It provides researchers with convenient access to a comprehensive collection of pathways from multiple sources represented in a common language. Access is via a web portal for browsing, query and download. Database providers can share their pathway data via a common repository and avoid duplication and reduce software development costs. Bioinformatics software developers can increase efficiency by sharing software components. Pathways include biochemical reactions, complex assembly, transport and catalysis events, and physical interactions involving proteins, DNA, RNA, small molecules and complexes.
For more information, please visit Pathway Commons web site.
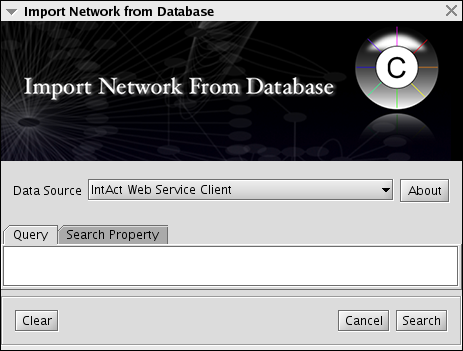
IntAct Client Plugin
General Information
Database Web Site |
|
WSDL |
http://www.ebi.ac.uk/intact/binary-search-ws/binarysearch?wsdl |
Data Type |
Protein-Protein interactions |
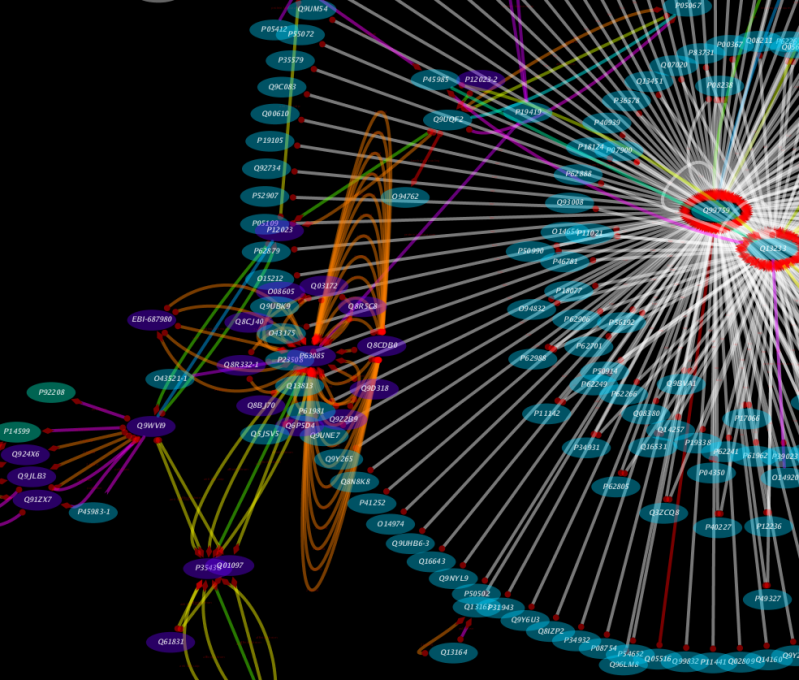
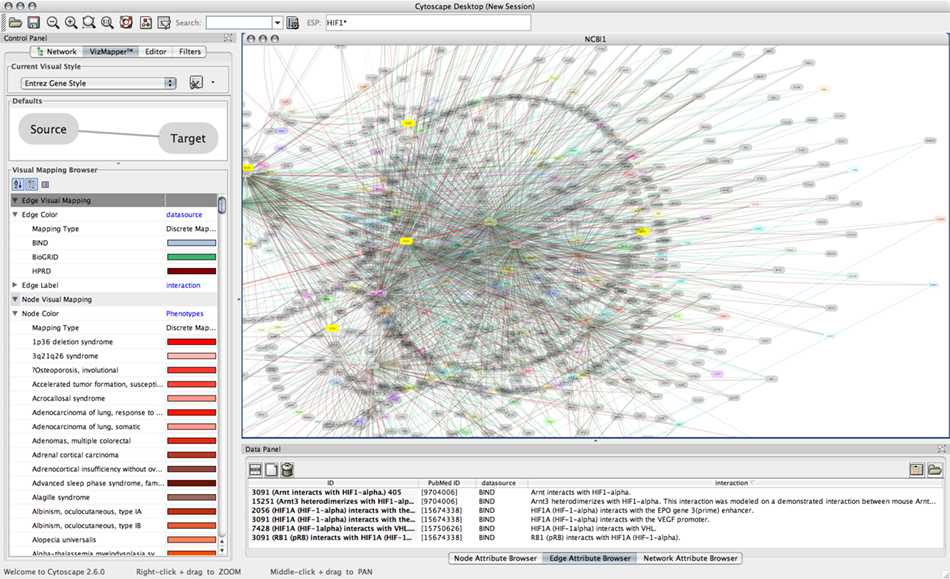
This client is for importing interaction data from IntAct. Because this is a network import clinet, the client will be automatically registered to the Unified Network Import user interface. Users can use Lucene sytax to search IntAct database. The network data includes a lot of node/edge information including:
- Aliases, Common Names, DB-Specific IDs
- GO Annotation
- Publication ID
- Interaction Detection Method
- Experimental Role (prey/bait)
The picture above is an example visualization of interaction data from IntAct. Visualization is based on attributes imported with the network.
How to Use
If a web service client is used for network import, it will be registered in the Unified Network Import User Interface:
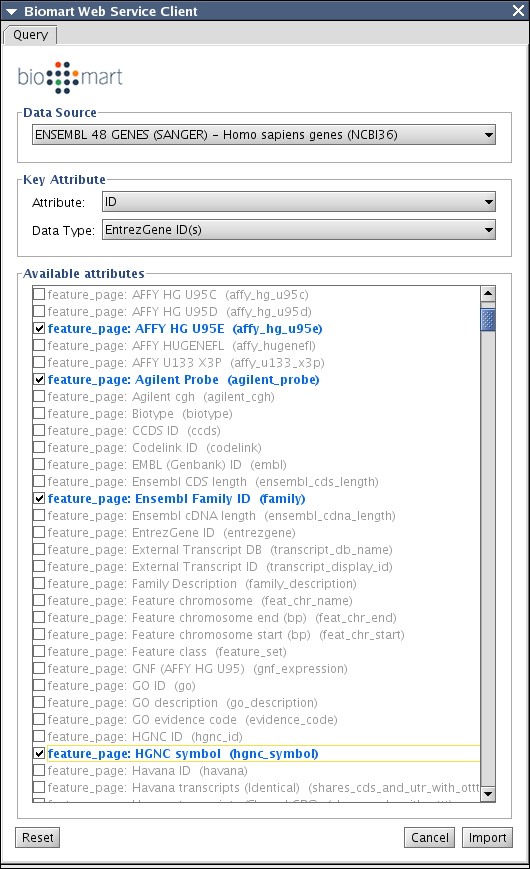
Biomart Client Plugin
Database Web Site |
|
WSDL |
N/A |
Data Type |
General Annotations |
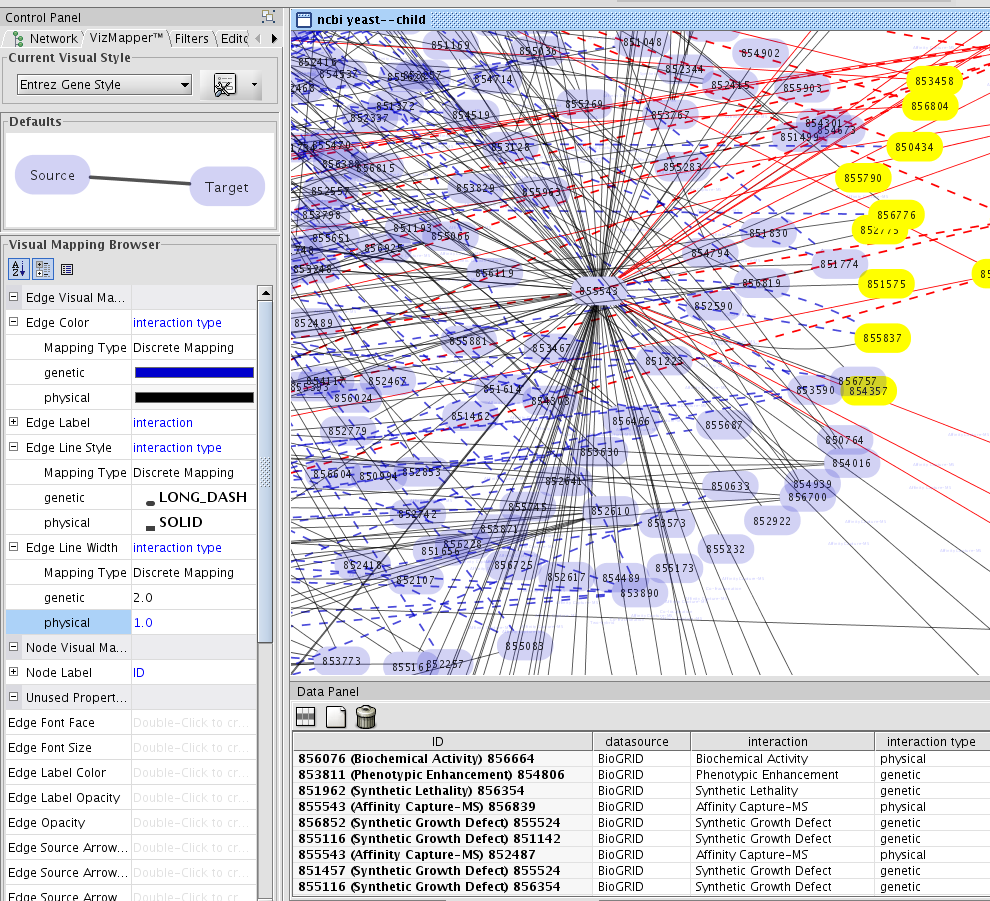
NCBI Entrez Client Plugin
Database Web Site |
|
WSDL |
|
Data Type |
General Annotations and Protein-Protein Interactions |
Overview
NCBI Entrez Client is the NCBI eUtils wrapped with Cytoscape WebServiceClient interface. Programatically, plugin developers can access full function set of eUtils. For users, this plugin have two functions:
Import network from Interactions section in Entrez Gene
- Import annotations from Entrez Gene
For each entry in Entrez Gene, it has both networks and annotations. This client can import both of these from the GUI above.
How to use
- Network Import:
File-->Import-->Network from web services...
For datasource, select NCBI Entrez Eutilities Web Service Client
Type free keyword (please read this page for search examples) and press Search
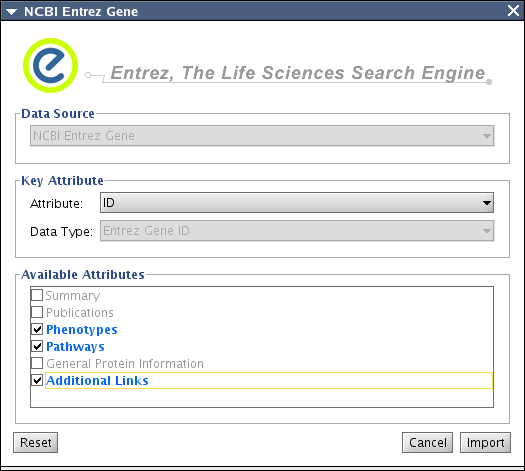
- Attribute Import:
File-->Import-->Import Attributes from NCBI Entrez Gene...
Select key attibute. This should be Entrez Gene ID. If the network node IDs are Entrez Gene ID, just select ID, otherwise, you need to import Entrez Gene ID somewhere. (And this is a good use case of BioMart client!)
- Select annotation category
Press Import
Note
Unfoetunatelly, NCBI service returns entire annotation for a query. This means even if you want to import only Official Name for your nodes in your network, this client needs to get the entire information for the node (gene/protein). Due to this issue, data import takes much longer than IntAct or BioMart client. Please be patient...
PICR Client Plugin
General Information
Database Web Site |
|
WSDL |
http://www.ebi.ac.uk/intact/binary-search-ws/binarysearch?wsdl |
Data Type |
Protein ID mapping |
Overview
- You can import various kinds of ID sets by this plugin. This plugin uses REST technology instead of SOAP.
If you have any questions, please email kono at ucsd.edu
Keiichiro Ono