Sample Workflows Using Web Service Clients
This is a simple example workflow with web service clients.
Prerequisites
- Cytoscape version 2.6.0 or later
- High-Speed Network Connection
- Plugins
- NCBIClient
- NCBIEntrezGeneUserInterface
BiomartClient
BiomartUserInterface
IntActClient
EnhancedSearch (optional)
Workflow 1: Start from Keyword
File-->Import-->Import network from web services...
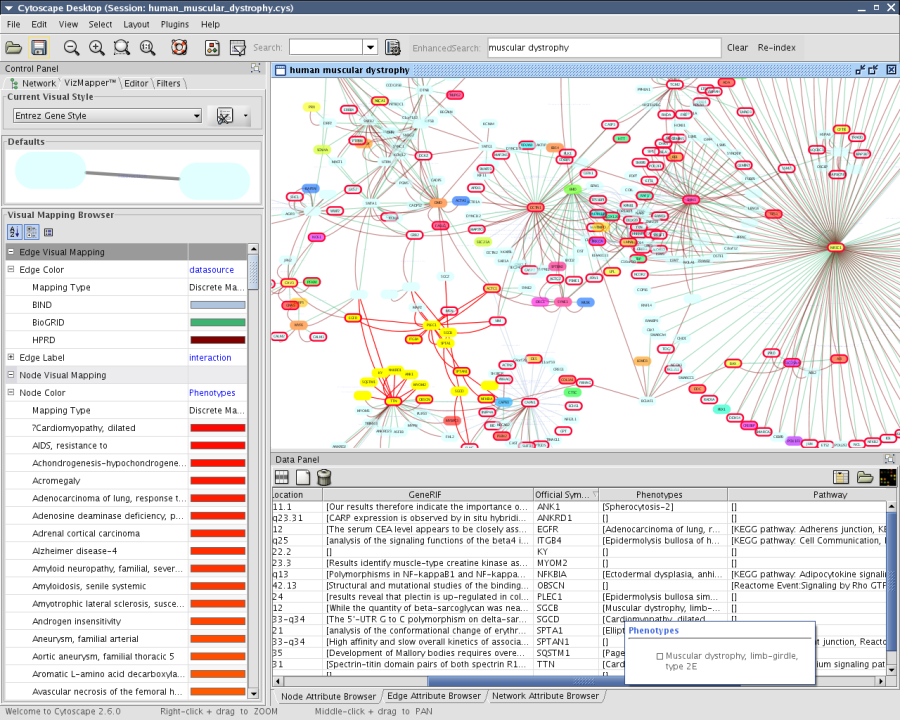
Select NCBI as datasource and type human muscular dystrophy as search keyword
Press Search. Number of candidates will be displayed. Then press OK. This process takes a long time.
File-->Import-->Import Attributes from NCBI Entrez Gene...
Select ID as key attribute and check all available annotations
Press Import. Again, this process takes a long time depends on network connection.
- Based on the attributes imported, visualize the network. (Example below: edge color is mapped to data source of the interaction, and phynotype is mapped to node color. Red border shows the node is on a known KEGG/Reactome pathway.)
'File-->Import-->Import attributes from BioMart...
Set ID as key attribute, and select the type to Entrez Gene ID
Select ENSEMBL 48 GENES (SANGER) - Homo sapiense genes (NCBI36) as data source
Check several attributes you want to import and then press Import
New attributes will be imported. Once you get new annotations, you can go to the datasource web page by Linkout feature.
Workflow 2: Start from List of IDs
Workflow 3: Start from User's Network