RFC Name : Cytoscape Data Integration |
Editor(s): Sarah Killcoyne, Alex Pico |
Status: Open for Comments |
<<TableOfContents: execution failed [Argument "maxdepth" must be an integer value, not "[2]"] (see also the log)>>
Proposal
Improve Cytoscape's data integration by creating a set of API's for some generic services that would be useful to the users and plugin developers. Create a registry and associated service taxonomy to allow users to search for and choose the service they are interested in using. Add a description of the services and API's to the wiki or manual for others to use when creating a new service.
Propsed Services
Synonym mapping
- Users request this frequently. If Cytoscape can provide the connectivity and the API of such a service others could create one that could be registered/discovered within Cytoscape allowing a user to pick the service they are interested in using.
- This would effectively deal with the requests we often get for a service based on the organism a researcher is working with (e.g. Yeast)
Particularly with regards to protein id services, I think this is entirely doable, the T1Dbase put together a database of mapping through various gene and protein ids, I think this would just break down into gene-gene mapping, gene-protein and protein-protein mapping services
Description of current ISB services: ISB_SynonymService.doc
Mammalian WSDL: ISB_Mammalian_WSDL.xml
Use cases specific to the service from Bader lab Bader Lab ID Mapping page:
Identifier translation. Some analysis methods require specific translations from one set of identifiers to another. For instance, our ‘activity centers’ analysis requires translation from protein or gene identifiers in a pathway database to Affymetrix probe set identifiers or other gene expression array platform identifiers. Finding a favorite gene name (e.g. Hugo or even lab-specific) is another type identifier translation.
Unification during dataset merging. During a merge operation e.g. of two protein-protein interaction datasets from independently created databases, it is vital to recognize that two protein objects, one from each data source, represent the same protein molecule, even if the protein objects don’t share any database accession numbers. Unification requires knowledge of record type e.g. you cannot reliably use a gene ID to unify proteins (mostly because splice variants exist).
Link out to related references. When presenting information about a protein to a user on a web page, it is useful to display links to related information about the protein, such as further information about the protein sequence and sequence feature annotations (e.g. in UniProt), Gene Ontology annotations, domains annotations (InterPro), etc.
Translating via orthology.Special case of identifier translation between species via orthology links. See HomoloGene.
- Example mapping plugin:
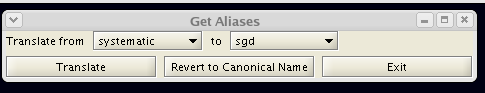
Network creation
Several plugins do this on their own already: cPath, AgilientLiteratureSearch, BioNetBuilder. If a common API could be arrived at for requesting/receiving network information the rest of the work can continue to be done by the respective servers instead of expecting plugins to build the networks as well.
- This would allow groups that currently offer their own data (e.g. Reactome) via web start to easily connect with Cytoscape users without having to create multiple web starts.
Use cases specific to the service:
Search for a network based on a set of identifiers or terms
Select the data to search, e.g. Reactome, Agilent or cPath
Retrieve network with annotation information. The data returned should able to encompass annotation/expression data along with the interaction data
Others?
- Any data tool that can be generic enough for a single API (WSDL)
Requirements
- A WSDL to be agreed upon for each service (see attached examples) as well as a standard representation (xml) for each requested object. REST may be the easiest to support and for others to implement but either SOAP or REST will use a WSDL. We could use a wiki to edit and maintain the WSDL over time (though this is in essence changing the API). These services would be tested for interoperability (e.g. document/literal WS-I basic profile).
- Taxonomy to describe services, with a manager to handle them (see registry). At the ISB we use three taxnomies (administrative, functional and descriptive) to describe different facets of a service.
- A registry to allow for search and discovery of services. This will allow users the flexibility to choose the service they wish to use (e.g. Yeast group will prefer to use the yeast synonym mapping service) See the following:
- S-Moby [attach the S-Moby PDF]
- UDDI [attach PDF from Oasis]
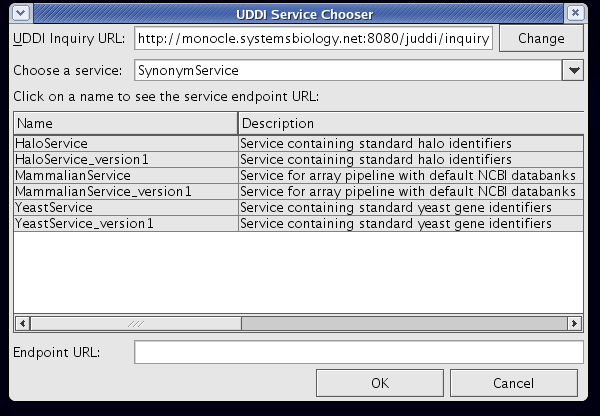
- ISB UDDI browser plugin:
Implementation Plan
These steps would be required for each service, the steps outlined below are specific to the Synonym Service because it is the first one we would like to implement. All time estimates are for 1 FTE. Time estimates will vary for the other services, though with experience implementing one any others should be simpler.
- Create WSDL for each service (start with id mapping as ISB and UCSF/Gladstone have working ones already)
- WSDL basically defines the API of a service
- Time: 2 weeks
- Deliverable - WSDL
- Create taxonomy to describe synonym services
- Time: 1 week
- Deliverable - Service taxonomy
- Create at least one service that implements the WSDL and taxonomy.
- ISB has a prototype in place that could be made public
- UCSF/Gladstone has ID mapping services it would like to reimplement as a web service
- Time: 2 weeks if we adopt our or another service, 16 weeks to create otherwise
- Deliverable - Usable services
- Choose and implement registry to locate services (see ISB’s UDDI browser plugin screenshots)
- There is already 3rd party software available for this used in the ISB browser
- Time: 2 weeks
- Deliverable - registry search backend
- Implement UI
- Time: 1 week
- Deliverable - UI
- Document WSDL, taxonomy and registry for other servers, unit tests
- Contact others who might be interested in implementing the service?
- Time: 3 weeks
- Deliverable - documentation and unit tests
Comments
How to Comment
Edit the page and add your comments under the provided header. By adding your ideas to the Wiki directly, we can more easily organize everyone's ideas, and keep clear records. Be sure to include today's date and your name for each comment. Try to keep your comments as concrete and constructive as possible. For example, if you find a part of the RFC makes no sense, please say so, but don't stop there. Take the extra step and propose alternatives.