Tutorial 3: Fetching External Data
Contents
If you have completed the Getting Started and Filters and Editor tutorials, this tutorial will help you get started with your own analysis by showing you some ways to fetch external data for Cytoscape. In this tutorial, you will:
- Download Cytoscape-formatted data from external servers including SGD and BIND.
- Fetch external data using the cPath plugin.
This tutorial and accompanying lectures were delivered at CSC, the Finnish IT center for science. The lecture slides of background material and an accompanying video presentation are available courtesy of the CSC at http://www.csc.fi/english/research/sciences/bioscience/Courses_and_events/cytoscape/index_html.
This tutorial features the following plugin (automatically included in Cytoscape 2.4 and up):
The cPath plugin from the Sander group at the Computational Biology Center at Memorial Sloan-Ketting Cancer Cente
Begin by (a) opening a second web browser window, and (b) clicking here: WEB START (approximate download size: 22 MB) This starts Cytoscape on your own computer, after downloading the program and annotations from our website.
External websites for SIF files
Here, we will explore some external data resources for Cytoscape.
Saccharomyces Genome Database (SGD)
This is a key resource for anyone who works with yeast! The SGD provides physical and genetic interactions, which may be downloaded into Cytoscape SIF format, as outlined below.
Go to http://db.yeastgenome.org/cgi-bin/batchDownload, and scroll down to the section labeled Step 1: Your Input.
- Under Enter Feature/Standard Gene names, enter a gene symbol such as PPA2.
- Under Step 2, under the section labeled Other data, check the boxes for physical and genetic interactions and click Submit.
- You will be redirected to a page labeled Download Data. Near the right side of this page is a link labeled Name of Downloadable File. Note the filename, and click on the link to download the file.
- If your system did not automatically uncompress the file during download, uncompress it.
- Load the uncompressed file into Cytoscape and apply your favorite layout algorithm.
- What sort of interactions are represented in this network? Physical or genetic? You can check by looking at the edge attributes under the Attribute Browser.
Biomolecular Interaction Network Database (BIND)
BIND is a central repository for protein interaction data, and represents a very useful resource for those interested in interaction networks. BIND is currently part of the BOND website, and can export data to the SIF format as described below.
Go to the BOND website (Biomolecular Object Network Databank) at http://bond.unleashedinformatics.com and log in. If you do not have a BOND account, create a free one and then log in.
- In the Search box, enter the name of your favorite protein (such as "TP53") and click on Go.
- This will take you to a results summary page. Click on the Interactions tab.
- On the left, you will see a link named "Interaction/Complex/Pathway Filters". Clicking on the (+) sign next to it will allow you to constrain what types of interactions to view. On the right, you will see three dropdown lists (View, Export Results, and Visualize Results).
- Select "Cytoscape SIF" from the Export Results list to download a file named searchresults.sif. This will allow you to load the network in your desktop version of Cytoscape.
Please note: This BIND service will eventually be superseded by a new Pathway Commons service, which is currently under development. Pathway Commons is an open access project that aims to provide a single access point for various existing public pathway databases (see http://www.pathguide.org/ for a comprehensive list), enabling efficient data storage, integration, and exchange.
cPath
Another useful resource for Cytoscape data is the cPath database and Cytoscape plugin. CPath is a free interaction database server package developed at Memorial Sloan Kettering Cancer Center to serve as a central clearinghouse for interaction and pathway data. By default the Cytoscape cPath plugin interfaces with a demo cPath database, hosted at MSKCC, with interaction data from the MINT and IntAct databases. Any lab working with interaction data from a variety of sources can install and populate a local copy of cPATH, and this will then allow them to fetch their data from one central repository. Here, we shall explore using the cPath plugin to fetch data from Cytoscape.
- If you have not already done so, start Cytoscape using the Webstart link at the top of this page. This will start an instance of Cytoscape 2.4 with the cPath plugin already loaded. This version connects to the cPath demo database at MSKCC.
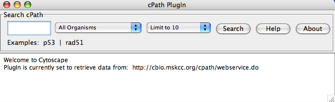
Go to File → New → Network → Construct network using cPath... . A window should appear, as shown below.
- In the dropdown list labeled All Organisms, select Homo Sapiens.
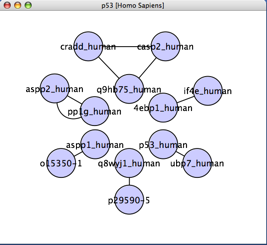
- In the box labeled Search cPath, enter P53 and click on the Search button. In your Cytoscape canvas, you should see a network similar to the one shown below (generated using the JGraph radial layout):
As you are probably aware by now, the number of interactions retrieved has a default limit of 10. However, you can change this via the dropdown list on the cPath plugin window. Please be kind to servers: When exploring a query, start with a low interaction limit. If the query proves useful, then increase your interaction limit gradually.
- You can use cPath to search on attributes including diseases (e.g. lymphoma) and biological processes (e.g. apoptosis). You can also combine search terms using the AND and OR operations (e.g p53 AND apoptosis). Experiment with this a bit. Remember to be kind to servers!
After users obtain network data from multiple sources, they often want to merge the separate networks into one network. This can be done with the Cytoscape Graph Merge plugin from the Ideker Lab at UCSD. For information on installing and using the plugin, please see the release notes for this plugin.
Congratulations! You have ventured past the world of Getting Started, and are now poised to start doing some serious systems biology! You will need your strength for this. Go get some lunch.
For comments or suggestions, please post to the cytoscape-discuss list.
Return to the Cytoscape introductory tutorials.