|
Size: 4443
Comment:
|
Size: 4417
Comment: converted to 1.6 markup
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 2: | Line 2: |
| Please write the use cases in terms of Cytoscape, meaning talk about node, edges, and attributes rather than talking about proteins and interactions. Instead, use specific biological discussions to illustrate instances of particular use cases. Try to be as general as possible. Try to see how your use case might match another use case that already exists. | |
| Line 3: | Line 4: |
| Please write the use cases in terms of Cytoscape, meaning talk about node, edges, and attributes rather than talking about proteins and interactions. Instead, use specific biological discussions to illustrate instances of particular use cases. Try to be as general as possible. Try to see how your use case might match another use case that already exists. |
== Use Case: Biochemical Reactions (HyperEdges) == A Cytoscape Edge typically connects two Nodes, which we commonly refer to as Source and Target Nodes. While this is sufficient for most cases, there are cases where a user may want to have Edges that connect more than two Nodes. One example of this is a ''Biochemical Reaction'', where an Edge represents a ''reaction'' that may have multiple ''substrates'', ''products'', and ''mediators''. An example of this is shown in the figure below, |
| Line 5: | Line 7: |
|
A Cytoscape Edge typically connects two Nodes, which we commonly refer to as Source and Target Nodes. While this is sufficient for most cases, there are cases where you may want to have Edges that connect more than two Nodes. One example of this is a ''Biochemical Reaction'', where an Edge represents a ''reaction'' that may have multiple ''substrates'', ''products'', and ''mediators''. An example of this is shown in the figure below, attachment:biochemcial_reaction_eg.png |
{{attachment:biochemcial_reaction_eg.png}} |
| Line 11: | Line 11: |
| There may also be cases in which you may want to connect one edge with another edge, for example to represent the activity of a ''molecular species'' that modulates the action of a ''catalyst''. This is illustrated in the figure below | There may also be cases in which a user may want to connect one edge with another edge, for example to represent the activity of a ''molecular species'' that modulates the action of a ''catalyst''. This is illustrated in the figure below |
| Line 13: | Line 13: |
| attachment:edge_connect_edge.png | {{attachment:edge_connect_edge.png}} |
| Line 17: | Line 17: |
| If you look closely at these two figures, you will see some small squares at the intersections of the edges. In Cytoscape, we refer to this small square as a !ConnectorNode. We also refer to the collection of edges and !ConnectorNode as a HyperEdge. | If you look closely at these two figures, you will see some small squares at the intersections of the edges. We refer to this small square as a !ConnectorNode. We also refer to the collection of edges and !ConnectorNode as a !HyperEdge. |
| Line 19: | Line 19: |
| Using Cytoscape's Editor and the ''Bio''''''Chemical''''''Reaction'' visual style, you can build and modify ''reactions'' by dragging and dropping shapes from the editor palette onto the main network view window. You can add ''products'', ''substrates'', and ''mediators'', to a ''reaction''. | The following two figures show examples of biochemical reactions -- Krebs Cycle and Glycolysis Reaction. |
| Line 21: | Line 21: |
| The figure below shows the Editor palette for the ''Bio''''''Chemical''''''Reaction'' visual style and the result of dragging/dropping the ''Add''''''Reaction'' shape from the palette onto the canvas. | The first figure below illustrates Glycolosis Reaction. Note the use of shared edges and multiple connections to the same Node within a !HyperEdge. |
| Line 23: | Line 23: |
|
attachment:hyperEdgeEditor.png The ''Add''''''Reaction'' palette shape serves as a template that enables you to add a connected set of nodes and edges to a network. You can change the names of the ''substrates'', ''products'', and ''mediators'' by editing them in Cytoscape's Attribute Browser. You can connect additional ''products'', ''substrates'', and ''products'' to a ''reaction'' by dragging their associated shapes from the palette and dropping them on the ''reaction's'' ''connector'' node. An example of a ''reaction'' with two ''products'' is shown in the figure below. attachment:hyperedge_two_products.png An example of a ''reaction'' with two ''products'' and two ''substrates'' is shown in the figure below. attachment:hyperedge_two_products_two_substrates.png You can connect a conventional Cytoscape node to a ''reaction'' by creating an edge from the conventional Cytoscape node to a !ConnectorNode (via drag/dropping an edge from the editor palette onto the node). Conversely, you can connect a ''reaction'' to a conventional Cytoscape node by creating an edge from the !ConnectorNode to the conventional Cytoscape node (via drag/dropping an edge from the editor palette onto the ''connector'' node). The following two figures show examples of BiochemicalReactions that you can build up using Cytoscape's editor. Once the !HyperEdgeEditor plugin is loaded, you can perform File->Import->!HyperEdge Sample Networks. There are two sample networks--Krebs Cycle and Glycolysis Reaction. The first figure below illustrates Glycolosis Reaction. Note the use of shared edges and multiple connections to the same Node within a !HyperEdge. attachment:glycolosis.png |
{{attachment:glycolosis.png}} |
| Line 45: | Line 27: |
| attachment:krebs_cycle.png | {{attachment:krebs_cycle.png}} |
| Line 47: | Line 29: |
|
=== A Closer Look === As was stated earlier, a ''Biochemical''''''Reaction'' is composed of a set of ''substrates'', ''products'', and ''mediators'', and a Hyper''''''Edge that connects them. Here are some basic definitions, plus some rules and constraints for working with Hyper''''''Edges: |
A detailed discussion of Hyper''''''Edge design issues can be found on the wiki page at Cytoscape_3.0/HyperEdges. == Use Case: Scientific Illustration == Cytoscape is used by researchers who need to publish and present their work. Often a user will want to include figures of Cytoscape networks in their publications and/or presentations. This requires presenting the Cytoscape network in a way that is intuitive to biologists. But Cytoscape networks are designed for computation, not presentation. The figure below shows a segment of a Cytoscape network for Wnt signalling, imported originally from BioPAX. {{attachment:wnt_biopax.png}} Contrast this with a manually illustrate Wnt signalling pathway, as shown in the figure below, derived from Wikipathways (http://www.wikipathways.org). {{attachment:wnt_wikipathway.png}} Note the use of annotational elements such as brackets, free text, and free-form sketched objects, such as the nuclear membrane shown in bottom left of the figure. Cytoscape 3.0 should support both styles of network views. To do this, there needs to be support for arbitrary graphical and textual annotation. Users should be able to define new shapes. We should define a set of constraints such that user-defined shapes that hold to those constraints can be supported by the underlying Cytoscape renderer. Users should also have control over how different shapes will overlap. To this end, we have begun to integrate the construct of multiple Layers into the Cytoscape canvas class. The figure below shows an initial implementation of an interface between Wiki''''''pathways and Cytoscape, written by Thomas Kelder at University of Maastricht, which imports a network written in the GPML markup format that underlies Wiki''''''pathways. Note the use of custom shapes, such as brackets, and free form text. {{attachment:wnt-gpml.png}} Note the use of custom shapes, such as brackets, and free form text. We should discuss the implications of implementing such constracts more directly in Cytoscape, from the perspectives of performance, expressiveness, and usability. |
Cytoscape 3.0 Use Cases
Please write the use cases in terms of Cytoscape, meaning talk about node, edges, and attributes rather than talking about proteins and interactions. Instead, use specific biological discussions to illustrate instances of particular use cases. Try to be as general as possible. Try to see how your use case might match another use case that already exists.
Use Case: Biochemical Reactions (HyperEdges)
A Cytoscape Edge typically connects two Nodes, which we commonly refer to as Source and Target Nodes. While this is sufficient for most cases, there are cases where a user may want to have Edges that connect more than two Nodes. One example of this is a Biochemical Reaction, where an Edge represents a reaction that may have multiple substrates, products, and mediators. An example of this is shown in the figure below,
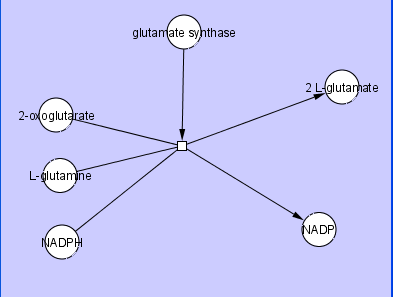
where L-glutamine and 2-oxoglutarate are substrates for the reaction, the catalyst glutamate substrate is a mediator for the reaction, 2 L-glutamate is a product of the reaction, the co-factor NADPH is a substrate for the reaction, and the co-factor NADP is a product of the reaction.
There may also be cases in which a user may want to connect one edge with another edge, for example to represent the activity of a molecular species that modulates the action of a catalyst. This is illustrated in the figure below
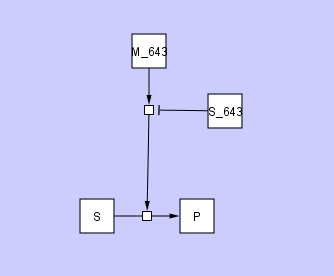
where molecular species S_643 inhibits the action of catalyst M_643.
If you look closely at these two figures, you will see some small squares at the intersections of the edges. We refer to this small square as a ConnectorNode. We also refer to the collection of edges and ConnectorNode as a HyperEdge.
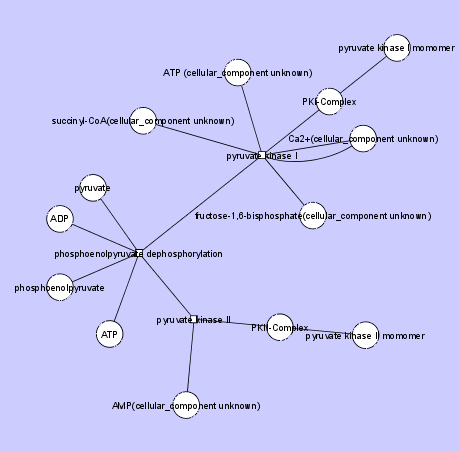
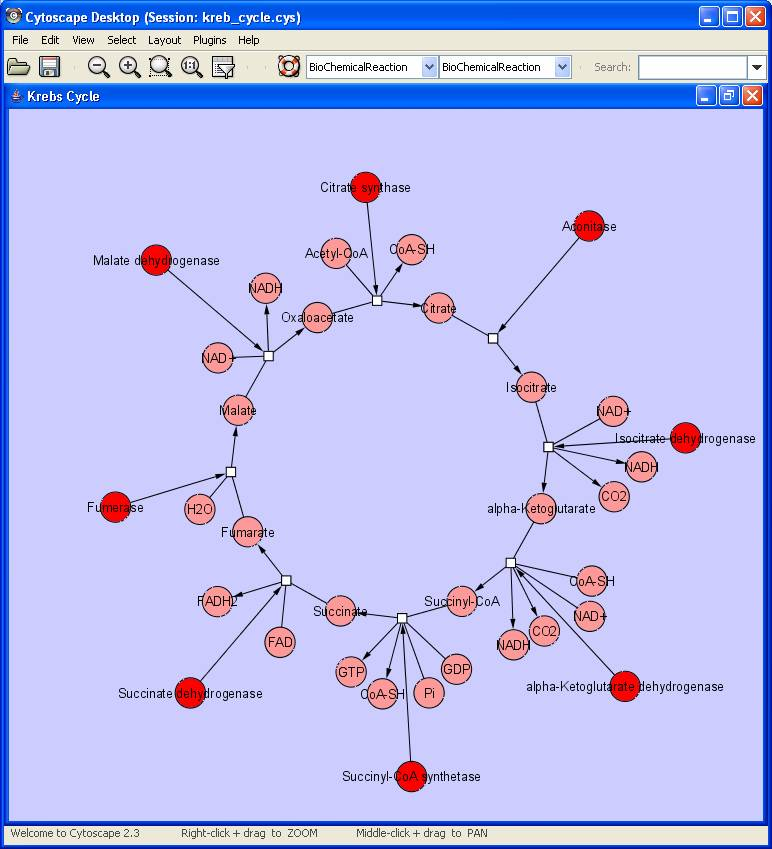
The following two figures show examples of biochemical reactions -- Krebs Cycle and Glycolysis Reaction.
The first figure below illustrates Glycolosis Reaction. Note the use of shared edges and multiple connections to the same Node within a HyperEdge.
The second example below uses a Circle Graph Layout to illustrate the Krebs Cycle.
A detailed discussion of HyperEdge design issues can be found on the wiki page at Cytoscape_3.0/HyperEdges.
Use Case: Scientific Illustration
Cytoscape is used by researchers who need to publish and present their work. Often a user will want to include figures of Cytoscape networks in their publications and/or presentations. This requires presenting the Cytoscape network in a way that is intuitive to biologists. But Cytoscape networks are designed for computation, not presentation. The figure below shows a segment of a Cytoscape network for Wnt signalling, imported originally from BioPAX.
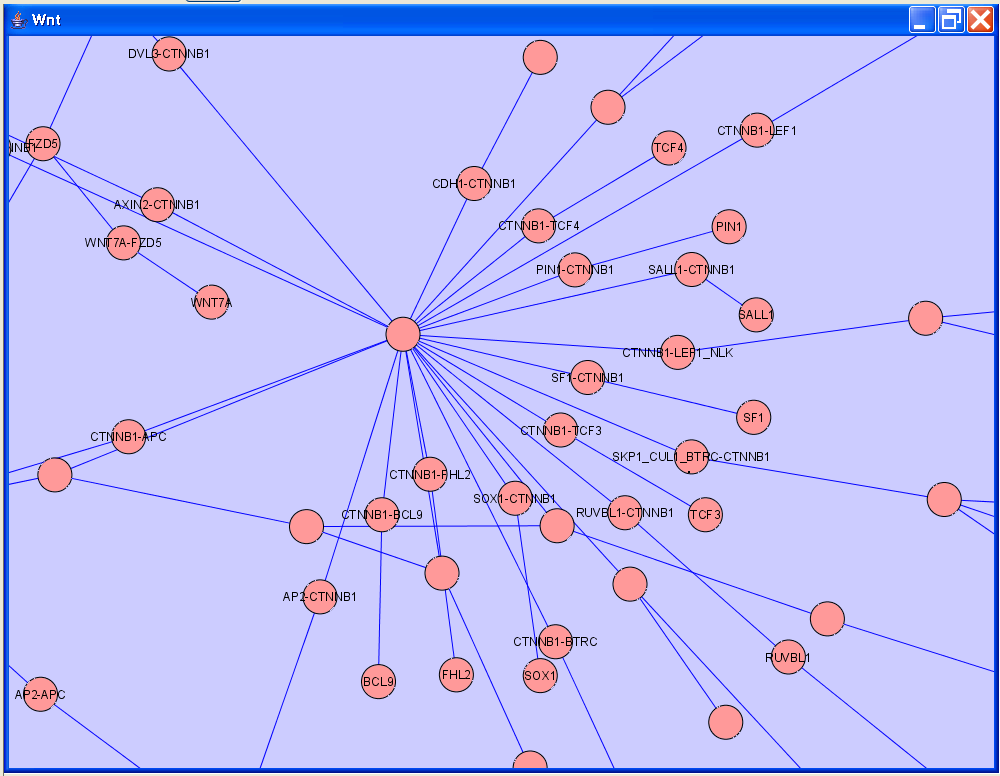
Contrast this with a manually illustrate Wnt signalling pathway, as shown in the figure below, derived from Wikipathways (http://www.wikipathways.org).
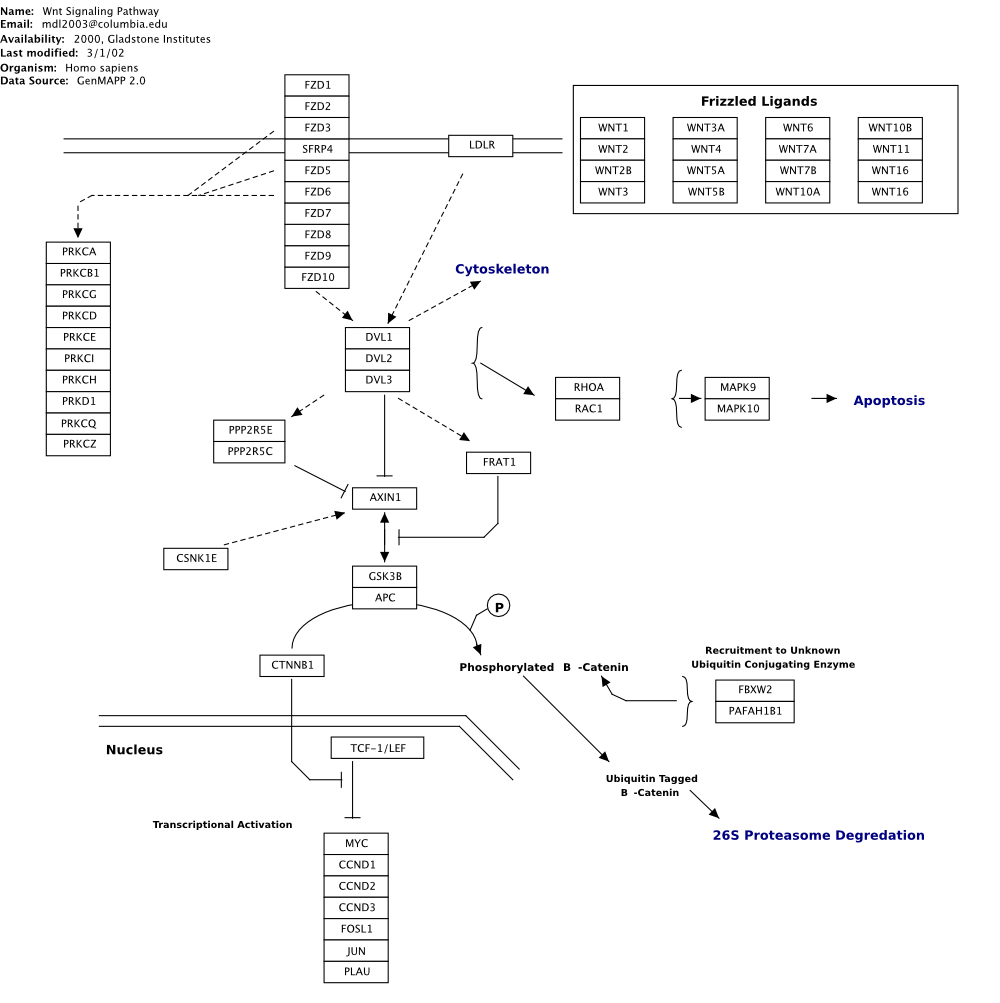
Note the use of annotational elements such as brackets, free text, and free-form sketched objects, such as the nuclear membrane shown in bottom left of the figure.
Cytoscape 3.0 should support both styles of network views. To do this, there needs to be support for arbitrary graphical and textual annotation. Users should be able to define new shapes. We should define a set of constraints such that user-defined shapes that hold to those constraints can be supported by the underlying Cytoscape renderer. Users should also have control over how different shapes will overlap. To this end, we have begun to integrate the construct of multiple Layers into the Cytoscape canvas class.
The figure below shows an initial implementation of an interface between Wikipathways and Cytoscape, written by Thomas Kelder at University of Maastricht, which imports a network written in the GPML markup format that underlies Wikipathways. Note the use of custom shapes, such as brackets, and free form text.
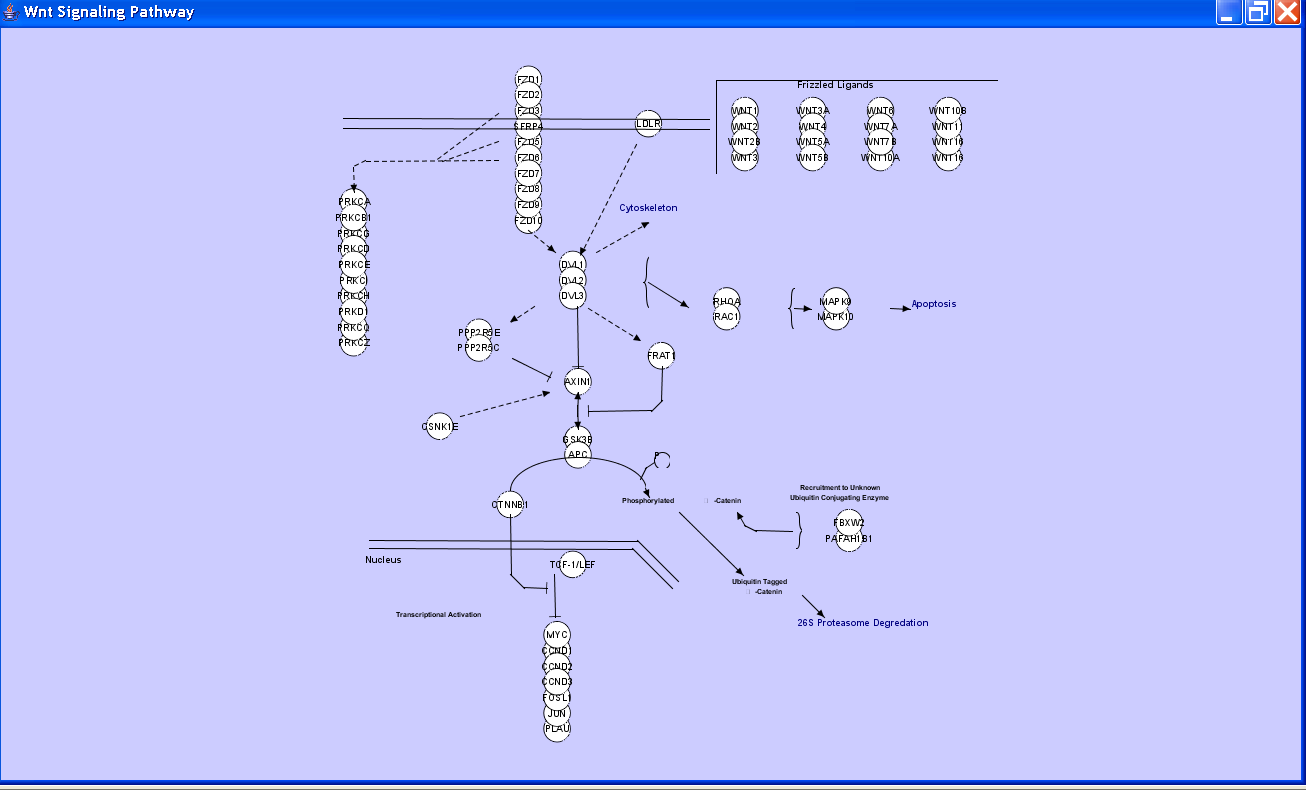
Note the use of custom shapes, such as brackets, and free form text. We should discuss the implications of implementing such constracts more directly in Cytoscape, from the perspectives of performance, expressiveness, and usability.