RFC66: Filter API |
Editor(s): Jason Montojo |
Date: 2010-11-09 |
Status: Open for comment until 2010-11-26 |
Proposal
A new filter API that retains the feature set of Cytoscape 2.X while adhering to the design philosophy of Cytoscape 3.0.
Background
Filters allow Cytoscape users to select a subset of nodes or edges from a network based on some condition. They can be named and saved for later use. Filters can also be applied to any network. The API should be general enough to allow the construction of filters that can check arbitrary conditions including, but not restricted to, node/edge/network attributes and graph topology. The API should also allow filters to be used through a scripting interface.
In Cytoscape 3.0 we need a clear separation between API and internal implementation. There was no such separation in 2.X. This means we need to formalize the abstractions for creating, registering, applying, combining, and (de)serializing filters.
Use Cases
The following use cases are currently supported by the filter implementation in 2.X. These should also be supported in 3.0.
- Create a new named filter using the Cytoscape GUI which selects node and edges based on conditions applied to a single attribute.
- Create a new named filter using the Cytoscape GUI which selects node and edges based on graph topology (e.g. node degree).
- Adjust the parameters of a named filter using the GUI.
- Use sliders to dynamically tune the parameters of a filter and show the resulting node/edge selection in real time.
- Apply a filter using the GUI.
- Compute the set-complement of a filter (i.e. invert the selection).
- Save filter definitions to a session file.
- Load filter definitions from a session file.
- Combine multiple filters into a single composite filter using set-union and set-intersection semantics.
- Programmatically create, configure and apply a filter to a network.
The following use cases are not supported by 2.X but should be possible in 3.0.
- Register a custom filter with Cytoscape and have it appear in the GUI.
- Write a script that uses a specific filter to select nodes and edges.
API
A proposed API which facilitates the use cases above is available for review here:
http://chianti.ucsd.edu/svn/core3/filter-api/trunk
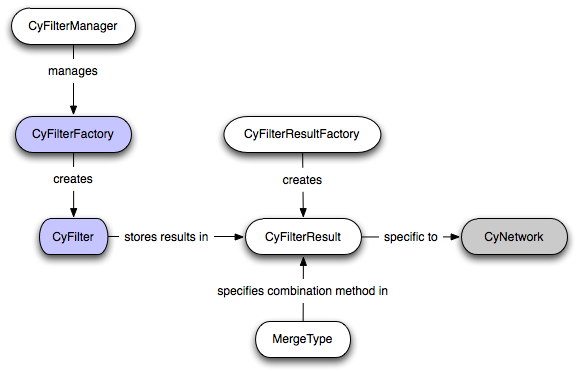
The following diagram illustrates how the various entities are related. Gray nodes are classes from external bundles. White nodes are classes provided by the API. Blue nodes are classes provided by filter implementors.
Performance Considerations
In order to maintain the same level of performance as 2.X, the implementation of this API should continue to use BitSets for tracking the nodes and edges selected by a filter. Using high performance primitive collections (e.g. long-array-backed HashSets) incur a 100% time penalty when merging, and reference-based collections incur a 1000% time penalty.
BitSet performance was observed to degrade when the total number of either the nodes or edges approached 10 million.
Comments
- Nothing yet.