|
Size: 3481
Comment:
|
← Revision 7 as of 2009-02-12 01:03:27
Size: 3808
Comment: converted to 1.6 markup
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 1: | Line 1: |
| ## page was renamed from ProcessStimulation | |
| Line 7: | Line 8: |
|
|| '''Use Case Name''' : ProcessStimulation || || '''For Feature''' : Self:MIMEditor || |
|| '''Use Case Name''' : ../ProcessStimulation || || '''For Feature''' : [[MIMEditor]] || |
| Line 16: | Line 17: |
| [[TableOfContents([2])]] | <<TableOfContents([2])>> |
| Line 35: | Line 36: |
| attachment:processstimulation1_061115_dwk.png | {{attachment:processstimulation1_061115_dwk.png}} |
| Line 39: | Line 40: |
| attachment:processstimulation2_061115_dwk.png | {{attachment:processstimulation2_061115_dwk.png}} |
| Line 45: | Line 46: |
| Assuming that this can use the same approach that is used for inhibition, it could be modeled as a Hyperedge containing node1: C (hyperedge attribute: process stimulation), node2: A, node3: B | Assuming that this can use the same approach that is used for ../ShowInhibition, it could be modeled as a Hyperedge containing node1: C (hyperedge attribute: process stimulation), node2: A, node3: B |
| Line 53: | Line 54: |
| This is of high importance. Stimulation of biological process by molecules that do not directly participate in the process is very common in biology. In some cases a process is contingent upon another reaction – in this case the relationship is defined as a Necessity. Other special casees include EnzymaticStimulation and EnzymaticStimulationInTrans, which have their own symbols. | This is of high importance. Stimulation of biological process by molecules that do not directly participate in the process is very common in biology. In some cases a process is contingent upon another reaction – in this case the relationship is defined as a ../ShowNecessity. Other special casees include ../EnzymaticStimulation and ../EnzymaticStimulationInTrans, which have their own symbols. |
| Line 59: | Line 60: |
| attachment:processstimulation3_061115_dwk.png |
{{attachment:processstimulation3_061115_dwk.png}} This example illustrates the concept of cooperative binding using the ../ProcessStimulation relationship. {{attachment:processstimulation4_061117_dwk.png}} |
| Line 65: | Line 70: |
| Shared MimEditorUseCaseComments | Shared ../MimEditorUseCaseComments |
| Line 68: | Line 73: |
| [[PageComment2]] |
---- AllanKuchinsky - 2006-11-26 04:25:13 should be handled in the same way as for ShowInhibition |
<<TableOfContents: execution failed [Argument "maxdepth" must be an integer value, not "[2]"] (see also the log)>>
Summary
A user wants specify that a species stimulates a reaction
Step-by-Step User Action
- User specifies a species to be the stimulator
- User specifies a reaction to be stimulated
- User specifies that there is a stimulation relationship
- User optionally specifies the evidence for the relationship
Visual Aides
The Kohn notation for process stimulation is the following:

An example using this notation follows:
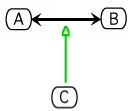
Requirements for Cytoscape
Assuming that this can use the same approach that is used for ../ShowInhibition, it could be modeled as a Hyperedge containing node1: C (hyperedge attribute: process stimulation), node2: A, node3: B
View: Use case for hyperedge view
Importance
This is of high importance. Stimulation of biological process by molecules that do not directly participate in the process is very common in biology. In some cases a process is contingent upon another reaction – in this case the relationship is defined as a ../ShowNecessity. Other special casees include ../EnzymaticStimulation and ../EnzymaticStimulationInTrans, which have their own symbols.
Variations
The stimulator does not necessarily have to be a molecular species; it can be a modified species or a process. In the example below, the phosphorylation of a molecule A enhances its ability to bind another molecule B:
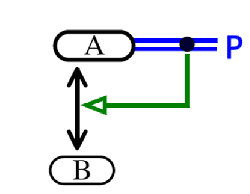
This example illustrates the concept of cooperative binding using the ../ProcessStimulation relationship.
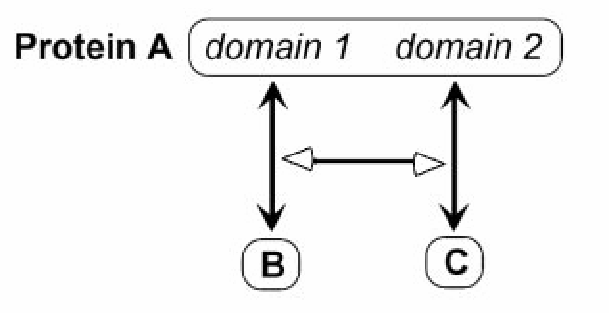
Other Examples
Comments
Shared ../MimEditorUseCaseComments
This could be modeled in BioPAX as either a Control (for catalysts) or a Biochemical Reaction.
AllanKuchinsky - 2006-11-26 04:25:13 should be handled in the same way as for ShowInhibition