<<TableOfContents: execution failed [Argument "maxdepth" must be an integer value, not "[2]"] (see also the log)>>
Summary
A user wants specify that a species is an enzyme that stimulates a reaction
Step-by-Step User Action
- User specifies a species to be the enzymatic stimulator
- User specifies a reaction to be stimulated
- User specifies that there is a stimulation relationship
- User optionally specifies the evidence for the relationship
Visual Aides
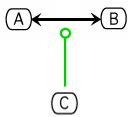
The Kohn notation for enzymatic stimulation is the following:

An example using this notation follows:
Requirements for Cytoscape
Assuming that this can use the same approach that is used for ../ShowInhibition, it could be modeled as a Hyperedge containing node1: C (hyperedge attribute: enzymatic stimulation), node2: A, node3: B
View: Use case for hyperedge view
Importance
This is of high importance, because it is very common in the maps.
Variations
When an enzyme stimulates two or more reactions, the enzyme reaction line branches and leads to enzyme reaction symbols at each target reaction.
TBD (Add a graphical example of this)
Other Examples
Comments
Shared ../MimEditorUseCaseComments
This is a special case of a ../ProcessStimulation. The enzyme binds to the complex as part of the reaction, and is unmodified after the reaction.
This could be modeled in BioPAX as a Catalysis object (e.g. Catalysis object with participants = a physicalInteraction object and a enzyme A, Controlled = the physicalInteraction object, Controller=A, Control-Type = Activation, and Direction = IRREVERSIBLE-LEFT-TO-RIGHT)
AllanKuchinsky - 2007-01-19 03:44:41
At the model level, this would be represented by ActivatingMediator nodes in HyperEdges.