|
Size: 8220
Comment:
|
← Revision 39 as of 2009-02-12 01:03:45
Size: 16986
Comment: converted to 1.6 markup
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 4: | Line 4: |
| Line 7: | Line 6: |
|
|| '''RFC Name''' : BioWebServiceConnectivity || '''Editor(s)''': KeiichiroOno || '''Status''': Open for Comment || [[TableOfContents([2])]] |
||'''RFC Name''' : BioWebServiceConnectivity ||'''Editor(s)''': KeiichiroOno ||'''Status''': Open for Comment || * Note 1: Some reference implementations are available now. Please visit SampleWebServiceClients page. * Note 2: [[WebServiceClientDevelopment| Guide for Web Service Client Developers]] is available. <<TableOfContents([2])>> |
| Line 13: | Line 14: |
|
(Still under construction!) These days, there are many public biological database web wervices such as [http://www.biomart.org/ Biomart], [http://www.ncbi.nlm.nih.gov/entrez/query/static/esoap_help.html NCBI], or [http://www.genome.jp/kegg/soap/ KEGG]. It is very useful for many users if Cytoscape have an unified access to those huge data resources through simple user interface. By this function, users can access virtually all kinds of annotations for the nodes and edges, including variety of ID's, sequences, descriptions, orthologs, pathway names which contains the nodes, etc. Also, this is a general solution for all types of node ID mapping problems Cytoscape currently have. In addition, users can access network (interaction) database web services through the same mechanism. From plugin writers' point of view, simplified API for accessing web services enables them to use a lot of remote functions just like other Cytoscape API. This means plugin writers can use web services as building blocks to build their workflow. |
These days, there are many public biological database web wervices such as [[http://www.biomart.org/|Biomart]], [[http://www.ncbi.nlm.nih.gov/entrez/query/static/esoap_help.html|NCBI]], or [[http://www.genome.jp/kegg/soap/|KEGG]]. It is very useful for many users if Cytoscape have an unified access to those huge data resources through simple user interface. By this function, users can access virtually all kinds of annotations for the nodes and edges, including variety of ID's, sequences, descriptions, orthologs, pathway names which contains the nodes, etc. Also, this is a general solution for all types of node ID mapping problems Cytoscape currently have. In addition, users can access network (interaction) database web services through the same mechanism. From plugin writers' point of view, simplified API for accessing web services enables them to use a lot of remote functions just like other Cytoscape API. This means plugin writers can use web services as building blocks to build their workflow. |
| Line 24: | Line 23: |
| * Implement an simple GUI for importing annotations and networks from the web services. | * Implement an simple GUI for importing annotations and networks from the web services. |
| Line 27: | Line 25: |
| Line 29: | Line 26: |
| * User loaded network with EntrezGene ID as node identifier into Cytoscape. He/she needs to import annotation files with Gene Symbol as the key. | * User loaded network with EntrezGene ID as node identifier into Cytoscape. He/she needs to import annotation files with Gene Symbol as the key. |
| Line 31: | Line 28: |
| * ''I want to expand existing network with known interaction data in [http://www.ebi.ac.uk/intact/site/index.jsf IntAct]'' | * ''I want to expand existing network with known interaction data in [[http://www.ebi.ac.uk/intact/site/index.jsf|IntAct]]'' |
| Line 34: | Line 31: |
| Line 36: | Line 32: |
| Each web service is a building block to make higher-level functions. This means plugin writers should be able to use registered web services as if they are local Java APIs. To achive this goal, we should be careful to separate UI from the web service clients. | Each web service is a building block to make higher-level functions. This means plugin writers should be able to use registered web services as if they are local Java APIs. To achive this goal, we should be careful to separate UI from the web service clients. |
| Line 40: | Line 36: |
| Line 41: | Line 38: |
|
* Maintaining list of web service clients. * GUI to view/edit properties of the clients. |
* Maintaining list of web service clients. * GUI to view/edit properties of the clients. |
| Line 44: | Line 41: |
| * Mapping fetched annotations onto !CyAttributes. | * Mapping fetched annotations onto !CyAttributes. |
| Line 46: | Line 43: |
|
* Implementing custom event and its listener only for !WebServiceClients * Higher-level functions, such as '''''importNetwork(Object query)''''' or '''''importAttribute(Object query)''''' will be called through this event handler. |
* Implementing custom event and its listener only for !WebServiceClients * Higher-level functions, such as '''''importNetwork(Object query)''' or '''''importAttribute(Object query)''''' will be called through this event handler. '' |
| Line 49: | Line 46: |
|
* Implement several web service clients for popular biological databases. * Each client is a jar file which contains properties and classes created from WSDL. They will be implemented as a regular Cytoscape plugins and loaded through Plugin Manager. * If WSDL is not available (like Biomart), we need to implement a simple stub to access the service. |
* Implement several web service clients for popular biological databases. * Each client is a jar file which contains properties and classes created from WSDL. They will be implemented as a regular Cytoscape plugins and loaded through Plugin Manager. * If WSDL is not available (like Biomart), we need to implement a simple stub to access the service. |
| Line 53: | Line 50: |
|
* GUI to use web services will be implemented as separate plugins. Simple GUI for importing attributes and networks will be provided as a part of core. |
* GUI to use web services will be implemented as separate plugins. Simple GUI for importing attributes and networks will be provided as a part of core. |
| Line 56: | Line 52: |
| Line 59: | Line 54: |
| * Browsing UDDI Registory (please read [[DataIntegration|this RFC]]) | |
| Line 61: | Line 56: |
|
* Pathway Commons / cPath integration to the new Network Import UI |
* Pathway Commons / cPath integration to the new simplifed Network Import GUI |
| Line 64: | Line 58: |
|
|
* Connection / integration to [[http://taverna.sourceforge.net/|Taverna]] or other workflow engins (like [[http://kepler-project.org/|Kepler]]). * More clients for other functions (analysis). * Use web services in batch (headless/daemon) mode analysis (3.0 or later). * Access from scripting languages (3.0) |
| Line 68: | Line 64: |
|
* [http://taverna.sourceforge.net/index.php?doc=services.html Biological Web Services] by myGrid/Taverna Project * [http://www.biomart.org/martservice.html Biomart] * [http://www.ncbi.nlm.nih.gov/entrez/query/static/esoap_help.html NCBI Entrez E-Utilities SOAP Service] * [http://www.ebi.ac.uk/~intact/devsite/modules/ws.html IntAct Web Services] * [http://www.ebi.ac.uk/uniprot/remotingAPI/ UniProtJAPI] * [http://www.genome.jp/kegg/soap/ KEGG API] * [http://www.reactome.org/cgi-bin/mart Reactome Mart Service] (Implemented with Biomart backend) |
* [[http://taverna.sourceforge.net/index.php?doc=services.html|Biological Web Services]] by myGrid/Taverna Project * [[http://www.biomart.org/martservice.html|Biomart]] * [[http://www.ncbi.nlm.nih.gov/entrez/query/static/esoap_help.html|NCBI Entrez E-Utilities SOAP Service]] * [[http://www.ebi.ac.uk/~intact/devsite/modules/ws.html|IntAct Web Services]] * [[http://www.ebi.ac.uk/uniprot/remotingAPI/|UniProtJAPI]] * [[http://www.genome.jp/kegg/soap/|KEGG API]] * [[http://www.reactome.org/cgi-bin/mart|Reactome Mart Service]] (Implemented by Biomart backend) |
| Line 78: | Line 72: |
|
* [http://ws.apache.org/axis/ Apache Axis] * [https://metro.dev.java.net/ GlassFish Metro Project] * [https://jax-ws.dev.java.net/ JAX-WS] (part of Metro project) |
* [[http://ws.apache.org/axis/|Apache Axis]] * [[https://metro.dev.java.net/|GlassFish Metro Project]] * [[https://jax-ws.dev.java.net/|JAX-WS]] (part of Metro project) |
| Line 83: | Line 76: |
| Line 85: | Line 77: |
|
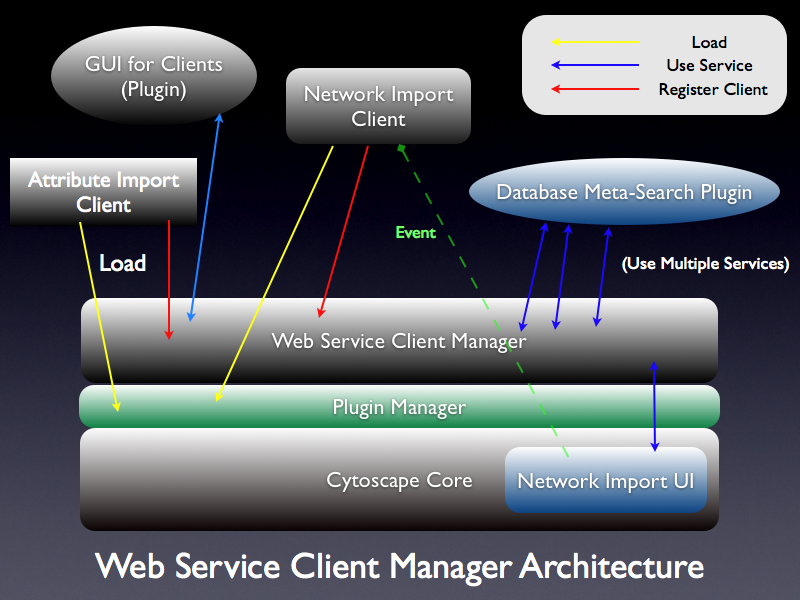
attachment:wsc_design.png Web Service Client Manager is a mechanism to manage list of clients loaded in Cytoscape. Usually, a Web Service Client (WSC) is created from a WSDL and packed as a jar file. The WSC jar file '''must''' contain classes implementing the following interfaces: |
. {{attachment:wsc_design.png}} '''Web Service Client Manager''' is a mechanism to manage list of clients loaded in Cytoscape. Usually, a Web Service Client (WSC) is created from a WSDL and packed as a jar file. The WSC jar file '''must''' contain classes implementing the following interfaces: |
| Line 91: | Line 82: |
|
This means each of WSC is a Cytoscape plugin. Therefore, all WSC will be loaded through Plugin Manager which is already in the core. Once WSC is loaded through Plugin Manager, WSC registers itself to Web Service Client Manager. Plugin writers can access all web services by using the following method: {{{ WebServiceClientManager.getClient(client_name); }}} |
This means each of WSC is a Cytoscape plugin. Therefore, all WSC will be loaded through Plugin Manager which is already in the core. Once WSC is loaded through Plugin Manager, WSC registers itself to Web Service Client Manager. Plugin writers can access all web services by using the following method: {{{ #!java WebServiceClient client = WebServiceClientManager.getClient(client_name); }}} In general, most of the methods required for clients are common to all. To avoid duplicate code, each web service client extends the superclass called ''WebServiceClientImpl''. Then, the actual client code will be similar to the following (this sample is for [[http://www.ebi.ac.uk/intact/site/index.jsf|IntAct]] network database): {{{ #!java public class IntactClient extends WebServiceClientImpl { private static final String DISPLAY_NAME = "IntAct Web Service Client"; private static final String CLIENT_ID = "intact"; // Client should be a singleton private static final WebServiceClient client; static { client = new IntactClient(); } /** * DOCUMENT ME! * * @return DOCUMENT ME! */ public static WebServiceClient getClient() { return client; } /** * Creates a new IntactClient object. */ private IntactClient() { super(CLIENT_ID, DISPLAY_NAME, new ClientType[] { ClientType.NETWORK }); stub = new BinarySearchServiceClient(); setProperty(); } /** * DOCUMENT ME! * * @param e DOCUMENT ME! */ @Override public void executeService(CyWebServiceEvent e) { if (e.getSource().equals(CLIENT_ID)) { if (e.getEventType().equals(WSEventType.IMPORT_NETWORK)) { importNetwork(e.getParameter(), null); } else if (e.getEventType().equals(WSEventType.EXPAND_NETWORK)) { importNetwork(e.getParameter(), Cytoscape.getCurrentNetwork()); } else if (e.getEventType().equals(WSEventType.SEARCH_DATABASE)) { search(e.getParameter().toString(), e); } } } private void importNetwork(Object searchResult, CyNetwork net) { // Actual network data import code will be placed here. } } }}} And the class to register this as a plugin will be the following: {{{ #!java public class IntactClientPlugin extends CytoscapePlugin { /** * Creates a new IntActClientPlugin object. */ public IntactClientPlugin() { // Register this client to the manager. WebServiceClientManager.registerClient(IntactClient.getClient()); } } }}} Plugin writers have two access method for these services: * Through the '''''Client Stub''''' * By calling '''''getStub()''' method, users can access all methods available from the service. Object type of the stub depends on the web service. For example, NCBI E-Utilities client returns '''''EUtilsServiceSoap''''' as the stub. If the client is registered properly, plugins can use it in the following way: '' {{{ #!java // Get client from the manager. WebServiceClient ncbiClient = WebServiceClientManager.getClient("ncbi"); // Then get the stub from the client. EUtilsServiceSoap ncbiStub = (EUtilsServiceSoap)ncbiClient.getStub(); // Use the stub EInfoResult res = ncbiStub.run_eInfo(new EInfoRequest()); }}} * Through CyWebServiceEvent * All clients implement !'''''CyWebServiceEventListener''''' and have mechanism to invoke method like ''importNetwork'' or ''importAttribute''. Plugin writers can access these methods by firing events. |
| Line 99: | Line 178: |
|
(Under construction) |
|
| Line 102: | Line 183: |
|
This event should contain the following information: * Type of event (IMPORT_NETWORK, IMPORT_ATTRIBUTE, EXECUTE_ANALYSIS, etc.) * Arguments (query command) |
|
| Line 103: | Line 188: |
|
|
|
| Line 106: | Line 189: |
|
Based on the architecture above, we need to design and implement several web service clients for popular biological databases. |
|
| Line 107: | Line 192: |
|
Web Service Clients use the following components in the core: * Client Property and Tunables * Network Import Panel * Attribute Import Panel |
Web Service Clients (WSC) use the following components in the core: 1. Client Property and Tunables * WSC should have a '''''WSCProperty''''' object which includes basic information of the client and property values which controls the behaviour of WSC. * Basic information * End Point (URI of the service) * Description * Example property values: * ''Search Mode'' - keyword or ID list. * ''Maximum Number of Result'' - Limit the number of search result (interactions / gene IDs) returned by the service. * These values will be managed through '''''Tunable''''' class. Currently it is used only by the Layout Manager, but will be generalized to edit all property values for WSC. * All of these properties are accessible from '''''WebServiceClientManagerDialog'''''. . {{attachment:wscm_dialog.png}} Left panel shows available (loaded) WSC and its category in the core. Right panel displays editable properties generated by ''Tunable''. 1. Network Import Panel . {{attachment:network_import_panel.png}} . This GUI component will be in the core for all network import WSCs. For simplicity, this panel accepts only two parameters: source database (source web service) and query string. Query string syntax depends on the service type. For example, IntAct accepts [[http://lucene.apache.org/java/docs/index.html|Apache Lucene]] syntax, and NCBI E-Utilities accepts list of !EntrezGene IDs ('''''run_eFetch''' method) or free keyword ('''''run_eSearch''''' method) as its parameter. In addition, we can pipeline search service and data fetch service(s) to accept keyword search (see the NCBI client example below). So we need a mechanism to switch the search mode for the clients (keyword search or data fetch based on ID list). Also, users may need to limit the number of interactions returned by service if the workstation's memory is limited. All of these will be encoded as properties and accessible from '''Option''' button. It just displays '''''WebServiceClientManagerDialog''''' discussed above. '' 1. Attribute Import Panel . {{attachment:attribute_import_panel.png}} . In many cases, attribute import client needs the following information to map attributes onto Cytoscape's data structure (CyAttributes): * Data Source - source database name * Attribute name used for mapping * Data type of the attribute - Official Symbol, EntrezGene ID, Ensembl Ac. Number, etc. * List of available attributes in the selected datasource. The GUI above will be in the core as the utility for attribute import clients. It takes icon and labels as its parameters. Checkbox list should be created by the client. (Example above is for [[http://beta.uniprot.org/|UniProt KB]]) |
| Line 116: | Line 221: |
|
attachment:biomartidmapping1.png Biomart client mainly used as ID mapping. |
{{attachment:biomartidmapping1.png}} Biomart client will be used as main datasource for ID mapping. For simple UI, ''Filter'' is always created from list of ID / Attribute. Since Biomart does not have published WSDL, we will implement a simple stub to accept XML query as the parameter. |
| Line 122: | Line 226: |
| Recently, UniProt released UniProt Java Remoting API (UniProtJAPI). We will wrap this API with the WebServiceClient interface in Cytoscape. | Recently, UniProt released UniProt Java Remoting API (UniProtJAPI). We will wrap this API with the WebServiceClient interface in Cytoscape. |
| Line 125: | Line 229: |
|
Most usuful information in KEGG is its [[http://www.genome.jp/kegg/pathway.html|pathway database]]. Most of them are accessible through [[http://www.genome.jp/kegg/soap/|KEGG API]]. We can use it to annotate networks in Cytoscape by fetching information theorugh their API. Pathway import function is optional since their pathway is not a simple graph object with binary-relationships (edges). We need to consider how to interpret ''reaction'' edges in Cytoscape (maybe HyperEdge?). |
|
| Line 127: | Line 234: |
|
Network import clients may have the following private methods which will be called via CyWebServiceEvent handlers: 1. '''''importNetwork(Object query)''''' * Create network based on the return values from the web service. 1. '''''expandNetwork(Object query)''''' {{attachment:expand_network.png}} . Expand function extracts list of selected node IDs and send it to the web service as the query. This will be called from context menu item in the current network view. Once the menu item is selected, CyWebServiceEvent wil lbe fired and listeners (network import web service clients) catch the event to execute ''expandNetwork'' method. Example above shows how to expand interactions around P35580 (MYH10) by IntAct web service client. |
|
| Line 129: | Line 243: |
| Cytoscape accesses IntAct through '''''BinarySearchServiceClient'''''. It takes Lucene syntax for query. We can add keyword search by pipelining search result from UniProtJAPI. | |
| Line 131: | Line 246: |
|
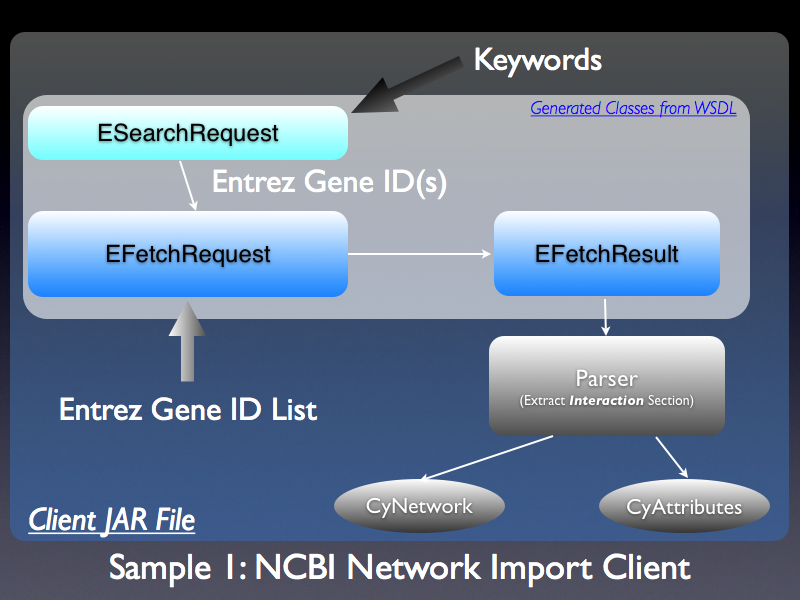
attachment:ncbi_network1.png NCBI Entrez database has a unified web service called [http://www.ncbi.nlm.nih.gov/entrez/query/static/esoap_java_help.html NCBI E-Utilities]. The [http://www.ncbi.nlm.nih.gov/sites/entrez?db=gene Entrez Gene] database will be the main resource for Cytoscape. This DB can be accessed through the E-Utilities web service. Users can use both keywords and list of Entrez Gene ID as the search term. Actual network data will be extracted form ''Interaction'' section of the returned data. Since we can extract interactions of one gene at a time, we need use Java SE 5's new Concurrency Framework to run multiple threads for fetching data efficiently. |
. {{attachment:ncbi_network1.png}} NCBI Entrez database has a unified web service called [[http://www.ncbi.nlm.nih.gov/entrez/query/static/esoap_java_help.html|NCBI E-Utilities]]. The [[http://www.ncbi.nlm.nih.gov/sites/entrez?db=gene|Entrez Gene]] database will be the main resource for Cytoscape. This DB can be accessed through the E-Utilities web service. Users can use both keywords and list of Entrez Gene ID as the search term. Actual network data will be extracted form ''Interaction'' section of the returned data. Since we can extract interactions of one gene at a time, we need use Java SE 5's new [[http://java.sun.com/j2se/1.5.0/docs/guide/concurrency/index.html|Concurrency Framework]] to run multiple threads for fetching data efficiently. |
| Line 136: | Line 250: |
| Line 138: | Line 251: |
| Line 140: | Line 252: |
| Edit the page and add your comments under the provided header. By adding your ideas to the Wiki directly, we can more easily organize everyone's ideas, and keep clear records. Be sure to include today's date and your name for each comment. '''Try to keep your comments as concrete and constructive as possible. For example, if you find a part of the RFC makes no sense, please say so, but don't stop there. Take the extra step and propose alternatives.''' | Edit the page and add your comments under the provided header. By adding your ideas to the Wiki directly, we can more easily organize everyone's ideas, and keep clear records. Be sure to include today's date and your name for each comment. '''Try to keep your comments as concrete and constructive as possible. For example, if you find a part of the RFC makes no sense, please say so, but don't stop there. Take the extra step and propose alternatives.''' |
| Line 144: | Line 256: |
| This RFC would seem to be very related to the [:DataIntegration: Data Integration] one. Might be useful to discuss this as part of a larger web services integration if we decide a larger, more general solution would be useful. |
This RFC would seem to be very related to the [[DataIntegration|Data Integration]] one. Might be useful to discuss this as part of a larger web services integration if we decide a larger, more general solution would be useful. AllanKuchinsky October 8, 2007 I would like to provide a 'preview window' for available pathways for a web service. I would like to be able to query a web service, such as PathwayCommons, for a list, preferably hierarchical, of available pathways, then present that list as a JTree (Windows Explorer-like interface) in a CytoPanel. When the user selects a pathway from the JTree, the pathway is imported via the Web service. |
RFC Name : BioWebServiceConnectivity |
Editor(s): KeiichiroOno |
Status: Open for Comment |
Note 1: Some reference implementations are available now. Please visit SampleWebServiceClients page.
Note 2: Guide for Web Service Client Developers is available.
<<TableOfContents: execution failed [Argument "maxdepth" must be an integer value, not "[2]"] (see also the log)>>
Proposal
These days, there are many public biological database web wervices such as Biomart, NCBI, or KEGG. It is very useful for many users if Cytoscape have an unified access to those huge data resources through simple user interface. By this function, users can access virtually all kinds of annotations for the nodes and edges, including variety of ID's, sequences, descriptions, orthologs, pathway names which contains the nodes, etc. Also, this is a general solution for all types of node ID mapping problems Cytoscape currently have. In addition, users can access network (interaction) database web services through the same mechanism.
From plugin writers' point of view, simplified API for accessing web services enables them to use a lot of remote functions just like other Cytoscape API. This means plugin writers can use web services as building blocks to build their workflow.
The goal of this project is the following:
Implement a mechanism to manage web service clients in the Cytoscape core (Web Service Client Manager).
- Design an interface which wraps each web service client.
- Design mechanism how web service clients communicate with the core (custom event handling).
- Implement an simple GUI for importing annotations and networks from the web services.
Biological Questions / Use Cases
User loaded network with EntrezGene ID as node identifier into Cytoscape. He/she needs to import annotation files with Gene Symbol as the key.
I need to check the orthologs for the genes in my network.
I want to expand existing network with known interaction data in IntAct
- Mapping known pathways onto network data in Cytoscape.
- Executing an analysis program running on remote server which is implemented as a web service.
General Notes
Each web service is a building block to make higher-level functions. This means plugin writers should be able to use registered web services as if they are local Java APIs. To achive this goal, we should be careful to separate UI from the web service clients.
Requirements
This project consists of five parts:
- Web Service Client Manager in the core
- Maintaining list of web service clients.
- GUI to view/edit properties of the clients.
- Attribute mapping module
Mapping fetched annotations onto CyAttributes.
- Event Handlers
Implementing custom event and its listener only for WebServiceClients
Higher-level functions, such as importNetwork(Object query) or importAttribute(Object query) will be called through this event handler.
- Sample Web Service Clients
- Implement several web service clients for popular biological databases.
- Each client is a jar file which contains properties and classes created from WSDL. They will be implemented as a regular Cytoscape plugins and loaded through Plugin Manager.
- If WSDL is not available (like Biomart), we need to implement a simple stub to access the service.
- GUI for each web services
- GUI to use web services will be implemented as separate plugins. Simple GUI for importing attributes and networks will be provided as a part of core.
Deferred Items
Open Issues
- Taxonomy of the services
Browsing UDDI Registory (please read this RFC)
Backward Compatibility
- Pathway Commons / cPath integration to the new simplifed Network Import GUI
Expected growth and plan for growth
Connection / integration to Taverna or other workflow engins (like Kepler).
- More clients for other functions (analysis).
- Use web services in batch (headless/daemon) mode analysis (3.0 or later).
- Access from scripting languages (3.0)
References
Biological Web Services
Biological Web Services by myGrid/Taverna Project
Reactome Mart Service (Implemented by Biomart backend)
Technologies Around Web Service
JAX-WS (part of Metro project)
Implementation Plan
Web Service Client Manager
Web Service Client Manager is a mechanism to manage list of clients loaded in Cytoscape. Usually, a Web Service Client (WSC) is created from a WSDL and packed as a jar file. The WSC jar file must contain classes implementing the following interfaces:
CytoscapePlugin
WebServiceClient
This means each of WSC is a Cytoscape plugin. Therefore, all WSC will be loaded through Plugin Manager which is already in the core. Once WSC is loaded through Plugin Manager, WSC registers itself to Web Service Client Manager. Plugin writers can access all web services by using the following method:
1 WebServiceClient client = WebServiceClientManager.getClient(client_name);
2
In general, most of the methods required for clients are common to all. To avoid duplicate code, each web service client extends the superclass called WebServiceClientImpl. Then, the actual client code will be similar to the following (this sample is for IntAct network database):
1 public class IntactClient extends WebServiceClientImpl {
2
3 private static final String DISPLAY_NAME = "IntAct Web Service Client";
4 private static final String CLIENT_ID = "intact";
5
6 // Client should be a singleton
7 private static final WebServiceClient client;
8 static {
9 client = new IntactClient();
10 }
11
12 /**
13 * DOCUMENT ME!
14 *
15 * @return DOCUMENT ME!
16 */
17 public static WebServiceClient getClient() {
18 return client;
19 }
20 /**
21 * Creates a new IntactClient object.
22 */
23 private IntactClient() {
24 super(CLIENT_ID, DISPLAY_NAME, new ClientType[] { ClientType.NETWORK });
25
26 stub = new BinarySearchServiceClient();
27
28 setProperty();
29 }
30
31 /**
32 * DOCUMENT ME!
33 *
34 * @param e DOCUMENT ME!
35 */
36 @Override
37 public void executeService(CyWebServiceEvent e) {
38 if (e.getSource().equals(CLIENT_ID)) {
39 if (e.getEventType().equals(WSEventType.IMPORT_NETWORK)) {
40 importNetwork(e.getParameter(), null);
41 } else if (e.getEventType().equals(WSEventType.EXPAND_NETWORK)) {
42 importNetwork(e.getParameter(), Cytoscape.getCurrentNetwork());
43 } else if (e.getEventType().equals(WSEventType.SEARCH_DATABASE)) {
44
45 search(e.getParameter().toString(), e);
46 }
47 }
48 }
49
50 private void importNetwork(Object searchResult, CyNetwork net) {
51 // Actual network data import code will be placed here.
52 }
53
54 }
55
And the class to register this as a plugin will be the following:
1 public class IntactClientPlugin extends CytoscapePlugin {
2 /**
3 * Creates a new IntActClientPlugin object.
4 */
5 public IntactClientPlugin() {
6 // Register this client to the manager.
7 WebServiceClientManager.registerClient(IntactClient.getClient());
8 }
9 }
10
Plugin writers have two access method for these services:
Through the Client Stub
By calling getStub() method, users can access all methods available from the service. Object type of the stub depends on the web service. For example, NCBI E-Utilities client returns EUtilsServiceSoap as the stub. If the client is registered properly, plugins can use it in the following way:
1 // Get client from the manager.
2 WebServiceClient ncbiClient = WebServiceClientManager.getClient("ncbi");
3 // Then get the stub from the client.
4 EUtilsServiceSoap ncbiStub = (EUtilsServiceSoap)ncbiClient.getStub();
5
6 // Use the stub
7 EInfoResult res = ncbiStub.run_eInfo(new EInfoRequest());
8
Through CyWebServiceEvent
All clients implement !CyWebServiceEventListener and have mechanism to invoke method like importNetwork or importAttribute. Plugin writers can access these methods by firing events.
Custom Event Model
(Under construction)
Instead of using standard PropertyChange events, core and clients sending custom events.
CyWebServiceEvent
This event should contain the following information:
- Type of event (IMPORT_NETWORK, IMPORT_ATTRIBUTE, EXECUTE_ANALYSIS, etc.)
- Arguments (query command)
CyWebServiceEventListener
Individual Client Design
Based on the architecture above, we need to design and implement several web service clients for popular biological databases.
Shared Components
Web Service Clients (WSC) use the following components in the core:
- Client Property and Tunables
WSC should have a WSCProperty object which includes basic information of the client and property values which controls the behaviour of WSC.
- Basic information
- End Point (URI of the service)
- Description
- Example property values:
Search Mode - keyword or ID list.
Maximum Number of Result - Limit the number of search result (interactions / gene IDs) returned by the service.
- Basic information
These values will be managed through Tunable class. Currently it is used only by the Layout Manager, but will be generalized to edit all property values for WSC.
All of these properties are accessible from WebServiceClientManagerDialog.
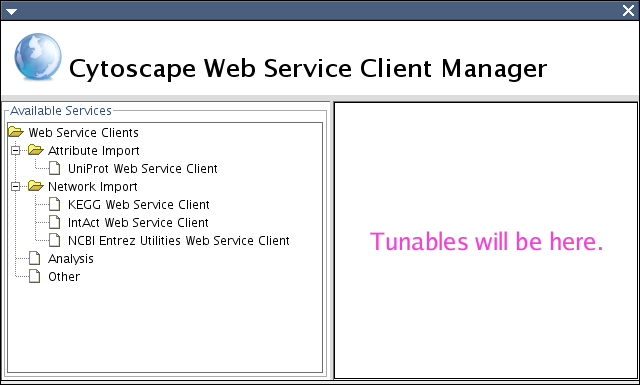 Left panel shows available (loaded) WSC and its category in the core. Right panel displays editable properties generated by Tunable.
Left panel shows available (loaded) WSC and its category in the core. Right panel displays editable properties generated by Tunable.
- Network Import Panel
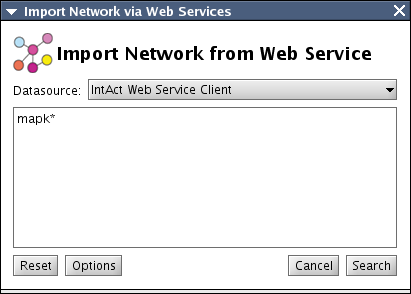
This GUI component will be in the core for all network import WSCs. For simplicity, this panel accepts only two parameters: source database (source web service) and query string. Query string syntax depends on the service type. For example, IntAct accepts Apache Lucene syntax, and NCBI E-Utilities accepts list of EntrezGene IDs (run_eFetch method) or free keyword (run_eSearch method) as its parameter. In addition, we can pipeline search service and data fetch service(s) to accept keyword search (see the NCBI client example below). So we need a mechanism to switch the search mode for the clients (keyword search or data fetch based on ID list). Also, users may need to limit the number of interactions returned by service if the workstation's memory is limited. All of these will be encoded as properties and accessible from Option button. It just displays WebServiceClientManagerDialog discussed above.
- Attribute Import Panel
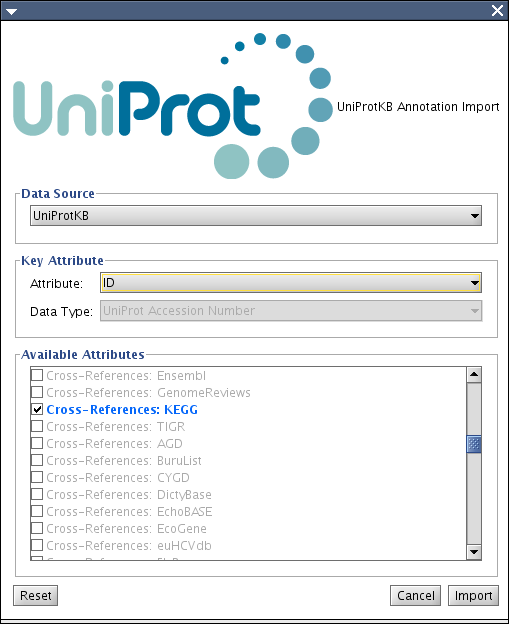
In many cases, attribute import client needs the following information to map attributes onto Cytoscape's data structure (CyAttributes):
- Data Source - source database name
- Attribute name used for mapping
Data type of the attribute - Official Symbol, EntrezGene ID, Ensembl Ac. Number, etc.
- List of available attributes in the selected datasource.
The GUI above will be in the core as the utility for attribute import clients. It takes icon and labels as its parameters. Checkbox list should be created by the client. (Example above is for UniProt KB)
ID Mapping and Attribute Import
Biomart
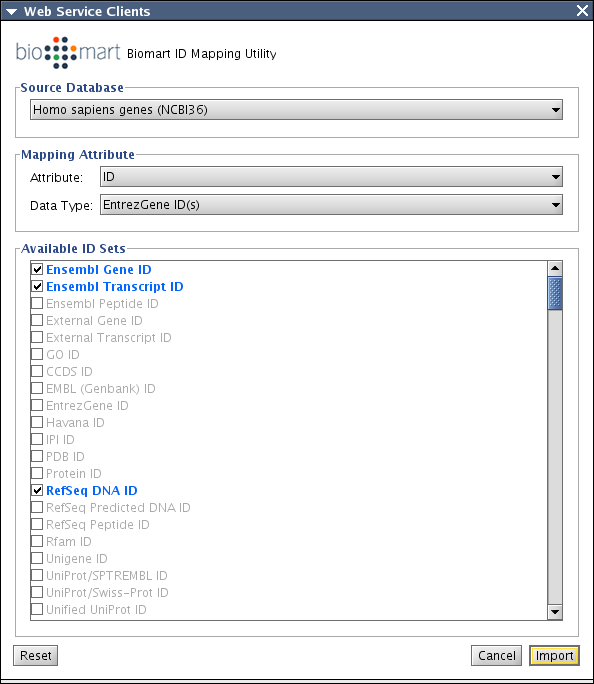
Biomart client will be used as main datasource for ID mapping. For simple UI, Filter is always created from list of ID / Attribute. Since Biomart does not have published WSDL, we will implement a simple stub to accept XML query as the parameter.
UniProt
Recently, UniProt released UniProt Java Remoting API (UniProtJAPI). We will wrap this API with the WebServiceClient interface in Cytoscape.
KEGG
Most usuful information in KEGG is its pathway database. Most of them are accessible through KEGG API. We can use it to annotate networks in Cytoscape by fetching information theorugh their API.
Pathway import function is optional since their pathway is not a simple graph object with binary-relationships (edges). We need to consider how to interpret reaction edges in Cytoscape (maybe HyperEdge?).
Network Import
Network import clients may have the following private methods which will be called via CyWebServiceEvent handlers:
importNetwork(Object query)
- Create network based on the return values from the web service.
expandNetwork(Object query)
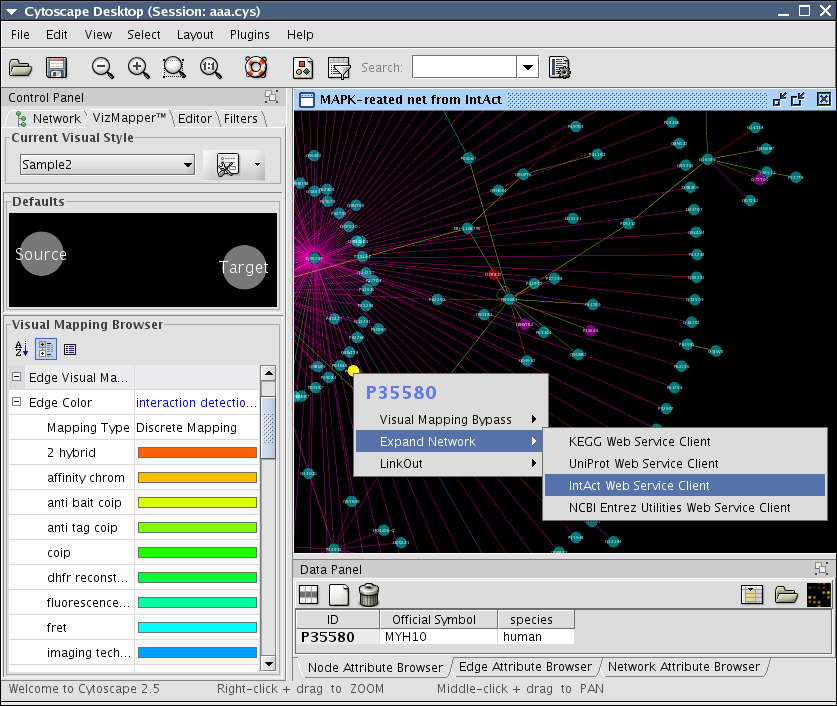
Expand function extracts list of selected node IDs and send it to the web service as the query. This will be called from context menu item in the current network view. Once the menu item is selected, CyWebServiceEvent wil lbe fired and listeners (network import web service clients) catch the event to execute expandNetwork method. Example above shows how to expand interactions around P35580 (MYH10) by IntAct web service client.
IntAct
Cytoscape accesses IntAct through BinarySearchServiceClient. It takes Lucene syntax for query. We can add keyword search by pipelining search result from UniProtJAPI.
NCBI
NCBI Entrez database has a unified web service called NCBI E-Utilities. The Entrez Gene database will be the main resource for Cytoscape. This DB can be accessed through the E-Utilities web service. Users can use both keywords and list of Entrez Gene ID as the search term. Actual network data will be extracted form Interaction section of the returned data. Since we can extract interactions of one gene at a time, we need use Java SE 5's new Concurrency Framework to run multiple threads for fetching data efficiently.
Comments
How to Comment
Edit the page and add your comments under the provided header. By adding your ideas to the Wiki directly, we can more easily organize everyone's ideas, and keep clear records. Be sure to include today's date and your name for each comment. Try to keep your comments as concrete and constructive as possible. For example, if you find a part of the RFC makes no sense, please say so, but don't stop there. Take the extra step and propose alternatives.
SarahKillcoyne August 16, 2007
This RFC would seem to be very related to the Data Integration one. Might be useful to discuss this as part of a larger web services integration if we decide a larger, more general solution would be useful.
AllanKuchinsky October 8, 2007
I would like to provide a 'preview window' for available pathways for a web service. I would like to be able to query a web service, such as PathwayCommons, for a list, preferably hierarchical, of available pathways, then present that list as a JTree (Windows Explorer-like interface) in a CytoPanel. When the user selects a pathway from the JTree, the pathway is imported via the Web service.