Use Case Name : Paralogs |
For Feature : Groups |
Editors: Kristina Hanspers |
<<TableOfContents: execution failed [Argument "maxdepth" must be an integer value, not "[2]"] (see also the log)>>
Summary
To represent two or more protein paralogs that can perform the same step in a pathway, we would like to group these. The set of proteins represent a logical OR, where either one of them alone can perform the reaction in question. To reduce complexity and enhance readability, it would be desirable to have these exhibit the properties of a group, in terms of collapsing etc, especially if there are many occurances of paralogs in one pathway.
Step-by-Step User Action
Creating a New Group of Paralogs
- Select two or more nodes
- Choose "Group Paralogs" or just “Group” (don’t know how specific this needs to be for the user) from a context menu, main menu or toolbar
- Automatically view collapsed view of group with default label (editable)
- Be able to expand group as vertically stacked set of nodes by a very simple mechanism (e.g., click on a plus/minus icon)
- perhaps other restricted views of the children will be allowed:
- horizontal stack
- block (e.g., 2x2, 3x4)
- overlapping "clump" of nodes packed into a some defined circular area
- perhaps other restricted views of the children will be allowed:
- Be able to destroy group
- Be able to expand-all or collapse-all groups in a given network
Loading a Network with Paralog Groups
- Same as 3-6 above
Visual Aides
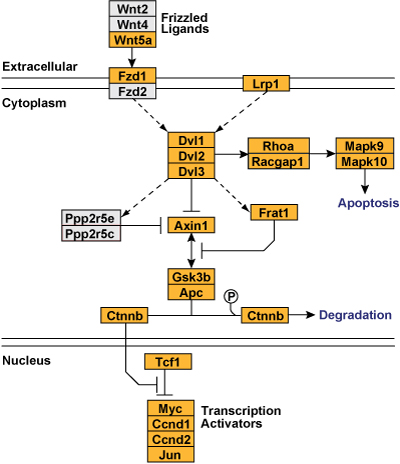
In this GenMAPP pathway, stacked gene objects help indicate complex and paralog groups. For example, Dvl paralogs.
Requirements for Cytoscape
- The Group API can handle this one in terms of create, expand, collapse and destroy
- A simple GUI for create, expand, collapse and destroy should be sufficient. Specific semantics for paralogs may not be necessary.
- Need to add simple layout/alignment algorithms for stacking children vertically in expanded view
- Need to support the storage of groups in a pathway file format. xGMML and GenMAPP's GPML?
Importance
Paralogs appear on many pathways, and being able to group them is important for readability of complex pathways.
Other Examples
- Simple, intuitive examples of expand and collapse can be found in directory tools/explorers that let you click on an icon (e.g., a triangle or plus/minus) to expand and collapse the view.
- Illustrator and other such programs have a grouping function that mimics the restriction on children once a group is formed. You cannot move the children relative to each other, for example. If you want to treat them independently, you have to ungroup.