|
Size: 20784
Comment:
|
Size: 21190
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 9: | Line 9: |
| * (!) Document revesion 1.4: Updated for Cytoscape IDE 0.1.0: KeiichiroOno (5/7/2009) | * (!) Document revesion 1.4: Updated for Cytoscape IDE 0.1.1. This is not finished yet. Will be ready 5/8/2009: KeiichiroOno (5/7/2009) |
| Line 88: | Line 88: |
|
* Test Platform 2 (Windows) __Still have some issues__ * Windows XP SP3 |
* Test Platform 2 (Windows) __Still have small issues__ * Windows XP SP3 32 Bit |
| Line 110: | Line 110: |
| 1. Click '''''Finish''''' | 1. Make sure '''''Check out All projects''''' checkbox is unchecked. Then click '''''Finish''''' |
| Line 115: | Line 115: |
|
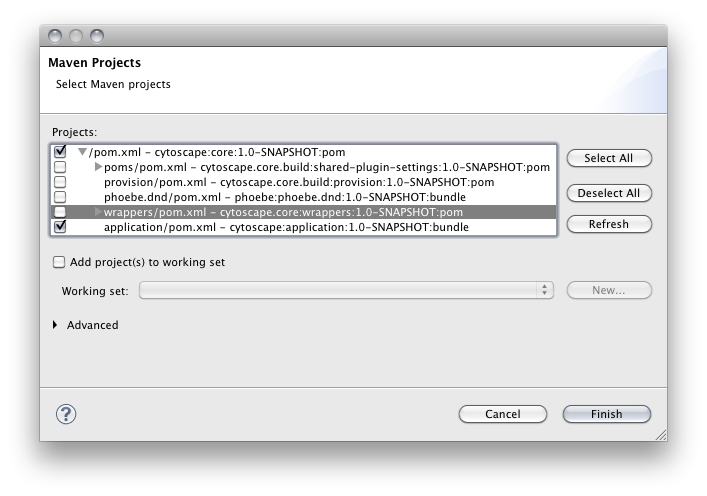
1. You will be asked to select maven projects you want to import. Select '''top-level pom''' and '''application''' ONLY. {{attachment:select_pom.png}} |
|
| Line 119: | Line 124: |
| == Step 3. Build Project and Run Cytoscape 3 on Felix == |
== Step 3. Build and Run Cytoscape 3 on OSGi Frameworks == {{attachment:cs3_nimbus1.png}} (Cytoscape 3 running on Windows XP + Java 6 update 13 with Nimbus L&F) |
Checkout, Compile, and Test Cytoscape 3 Core on Eclipse
Status
Document revesion 1.0 release: KeiichiroOno
Document revesion 1.1 release. Fixed minor issues based on feedback: KeiichiroOno
Document revesion 1.2 release. OSGi Project Nature section added: KeiichiroOno
Document revesion 1.2.1 release. VM Argument added for look & feel problem: KeiichiroOno
Document revesion 1.3.0 release. Tutorial is based on CytoscapeIDE: KeiichiroOno
Document revesion 1.3.1. Cytoscape IDE plugin is a bit out-of-date, so please install plugins one by one if you have any problems.: KeiichiroOno
 Document revesion 1.4: Updated for Cytoscape IDE 0.1.1. This is not finished yet. Will be ready 5/8/2009: KeiichiroOno (5/7/2009)
Document revesion 1.4: Updated for Cytoscape IDE 0.1.1. This is not finished yet. Will be ready 5/8/2009: KeiichiroOno (5/7/2009)
Contents
Introduction
This tutorial is for Java developers who wants to learn how to checkout code, compile, and test Cytoscape 3 with Eclipse PDE and other Eclipse plugins.
Tutorial
Step 1. Setup Your Environment
To develop Cytoscape 3 on Eclipse, you need to install several software packages.
1. Java Development Kit (JDK)
You need to install JDK version 5 or 6. Should work on both 32 bit and 64 bit versions. I tested this tutorial with the following JDK:
java version "1.6.0_13" Java(TM) SE Runtime Environment (build 1.6.0_13-b03) Java HotSpot(TM) Client VM (build 11.3-b02, mixed mode)
I strongly recommend to use Java 6 if it is available for your platform.
2. Eclipse PDE
This tutorial is based on the latest version of Eclipse 3.4, called Ganymede. Several distributions are available on Eclipse web site. In this tutorial, we use Eclipse for RCP/Plug-in Developers version 3.4. This distribution includes PDE (Plugin development environment). I recommend to install latest version of Eclipse PDE to a new directory on your machine. Otherwise, the screenshots may be different due to Eclipse plugins you have already installed on your machine.
You can install Eclipse by just extracting archive file (zip/gzip) to a directoly of your choice. By default, the eclipse directory should look like the following:
To keep your Eclipse up-to-date, you can update plugins by software updater (help-->software updates...)
For Windows Users: Set JDK JVM
Usually, Eclipse for Windows use JRE virtual machine instead of JDK's VM. To use JDK's VM, create a shortcut and edit the shortcut's property:
eclipse.exe -vm "c:¥path to jdk¥bin¥javaw.exe"
3. Cytoscape IDE
You can install all required Elcipse plugins for Cytoscape 3 development at once by installing Cytoscape IDE Plugin for Eclipse.
Select Help-->Software Updates...
Select Available Software tab
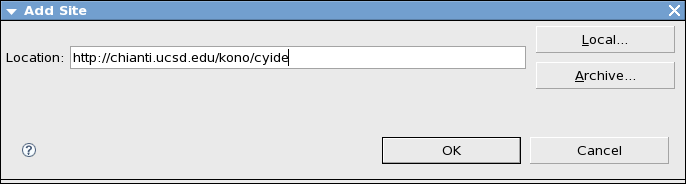
Click Add Site button and type http://chianti.ucsd.edu/kono/cyide in the Location text box. Then press OK.
Eclipse automatically scans the location and displays available plugins. You have to choose only one plugin, Cytoscape IDE for Eclipse
Press Install button and follow instructions. This process takes several minutes because it installes lots of plugins at once. When installation is complete, you will be asked to restart Eclipse. Press Yes and restart Eclipse.
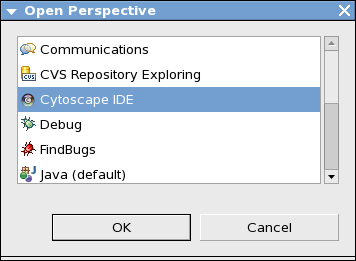
Select Window-->Open Perspective-->Other. Then you can find Cytoscape IDE as a new perspective. Select it and press OK
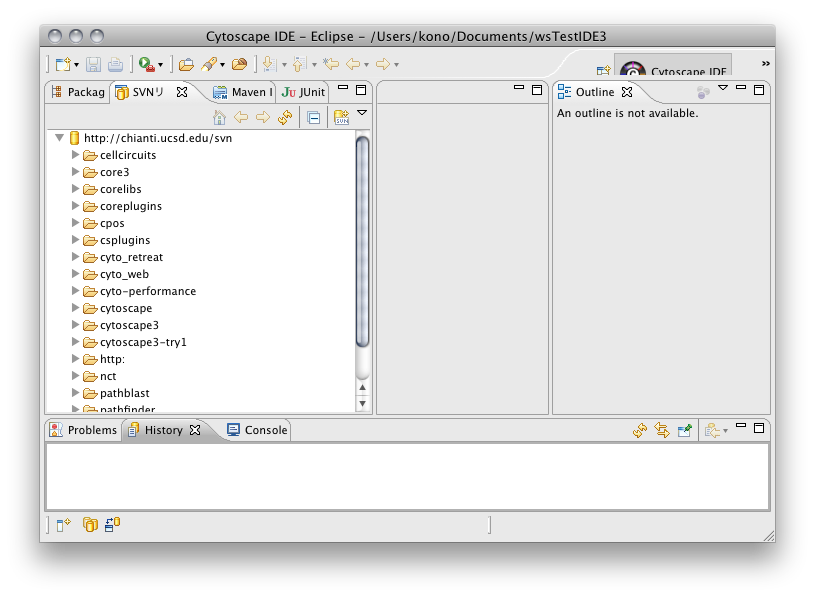
- If your workspace layout looks like the following, Eclipse is ready to develop Cytoscape 3.
Note: My Testing Environment
I've tested the following tutorial on these platforms:
- Common
- Eclipse PDE 3.4.2 Ganymede
Test Platform 1 (Unix) Confirmed to work
- RHEL 5 Desktop 64 bit
- Java SE 6 update 13 64 bit
Test Platform 2 (Windows) Still have small issues
- Windows XP SP3 32 Bit
JDK SE 6 update 13 Important Note: You should use JDK, not JRE.
- Test Platform 3 (Mac)
Step 2. Checkout and Create Cytoscape 3 Eclipse Project
In this section, you will learn how to create an Eclipse project from the fresh Cytoscape 3 checkout.
Checkout Cytoscape 3 Trunk
- Start Eclipse. I recommend to use an empty, new workspace only for Cytoscape 3 project since it creates tons of new bundle projects in your workspace. Make sure Cytoscape IDE perspective is selected.
 Turn off automatic build. (uncheck Project > Build Automatically)
Turn off automatic build. (uncheck Project > Build Automatically) Select SVN Repository tab. Cytoscape subversion repository will appear automatically.
If you are a core developer, use svn+ssh://grenache.ucsd.edu/common/svn. You need id and pssword. Otherwise, use http://chianti.ucsd.edu/svn. You can checkout the project from this URL, but cannot checkin.
The Cytoscape 3 repository is cytoscape3/trunk.
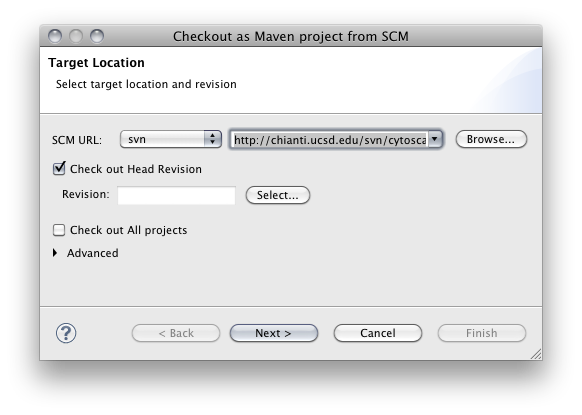
From SVN Repository window, right-click cytoscape3 > trunk and select Check out as Maven project...
Select Source Code Management (SCM) system type to svn and make sure the URL is set to http://chianti.ucsd.edu/svn/cytoscape3/trunk (for core developers, svn+ssh://grenache.ucsd.edu/common/svn/cytoscape3/trunk).
Make sure Check out All projects checkbox is unchecked. Then click Finish
You will be asked to select maven projects you want to import. Select top-level pom and application ONLY.
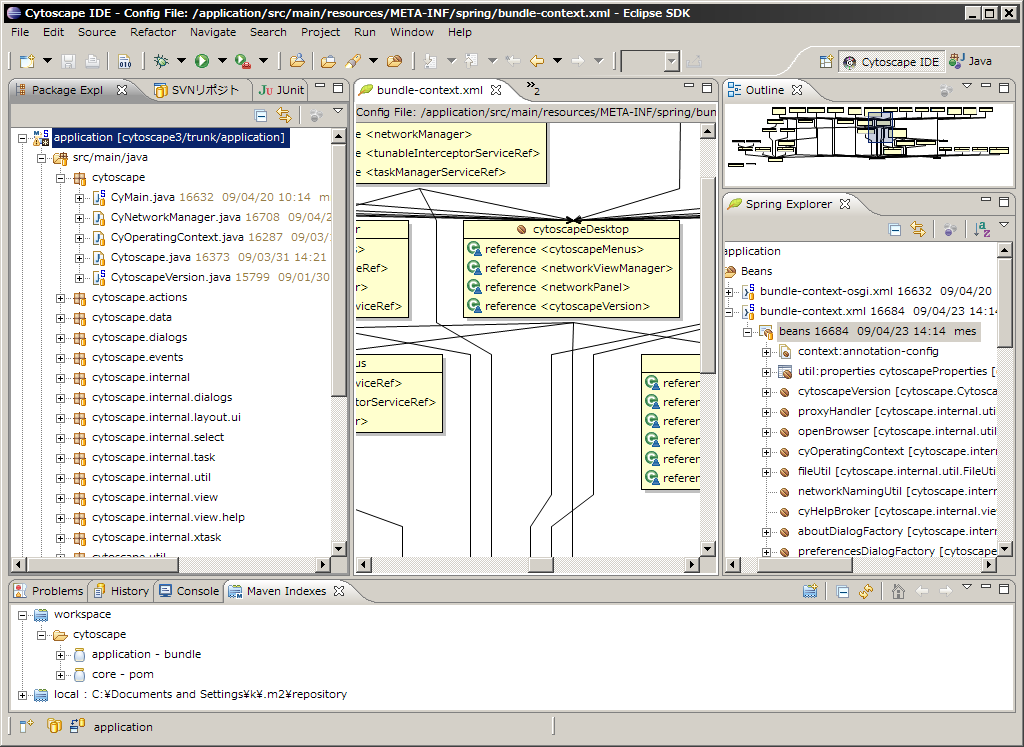
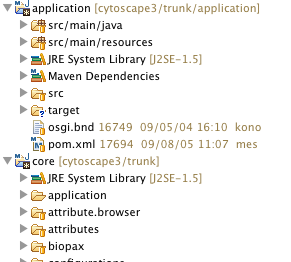
Select Package Explorer tab. Your Eclipse workspace looks like the following:
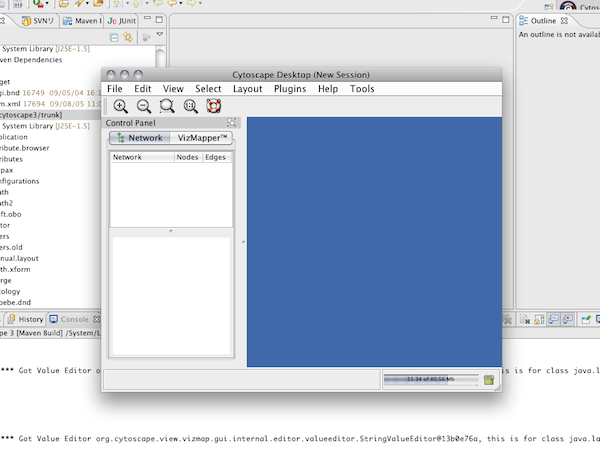
Step 3. Build and Run Cytoscape 3 on OSGi Frameworks
 (Cytoscape 3 running on Windows XP + Java 6 update 13 with Nimbus L&F)
(Cytoscape 3 running on Windows XP + Java 6 update 13 with Nimbus L&F)
First, let's build and run Cytoscape 3 from Eclipse workspace.
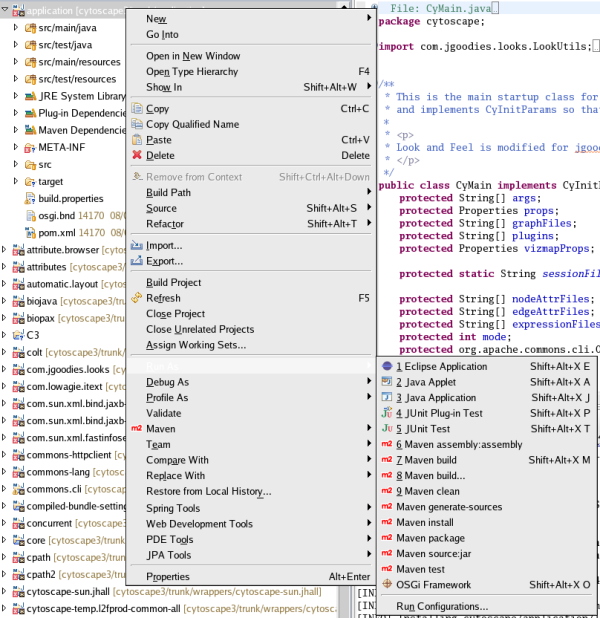
Select core project on the workspace. Right click and select Run As > Maven install. When you select this command, Eclipse builds and installs Cytoscape 3 bundles to your local maven repository ($HOME/.m2). It takes several minutes to complete (depends on your machine power).
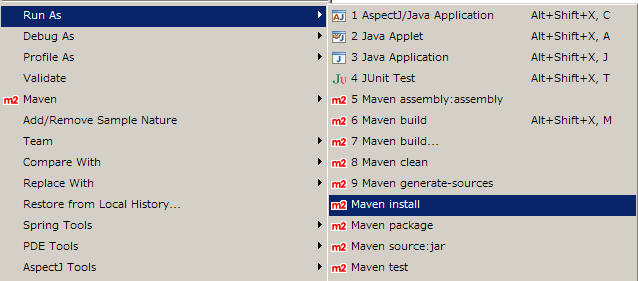
- If you cannot complete this process due to failing tests, try the following:
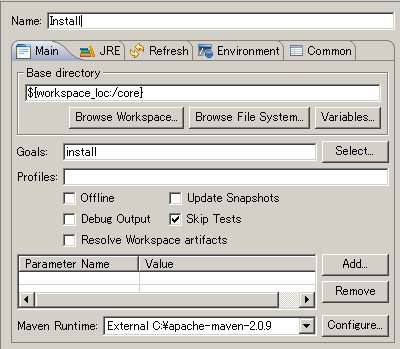
Select Run > Run Configurations
- Right-click Maven Build and create new setting
Set core as the base directory
Set install for goal and check Skip Tests
Apply and Run
This is a workaround for failing tests in Windows environment. We are working on this issue and once it is solved, you can call install goal from right-click menu.
- If you cannot complete this process due to failing tests, try the following:
Select Run → Run Configurations
Create new Maven Build and name it
Set the Base directory to core bundle
Set the goal to pax:provision
- Apply and Run. You can use Felix Shell commands on the console.
- Quit Cytoscape
Add OSGi Project Nature
At this point, each project in your workspace is defined as Maven Project and Eclipse can handle maven-specific features including dependency management and maven style source code directory structure. You can also add one more project nature to view/edit OSGi metadata file MANIFEST.MF by adding OSGi project nature.
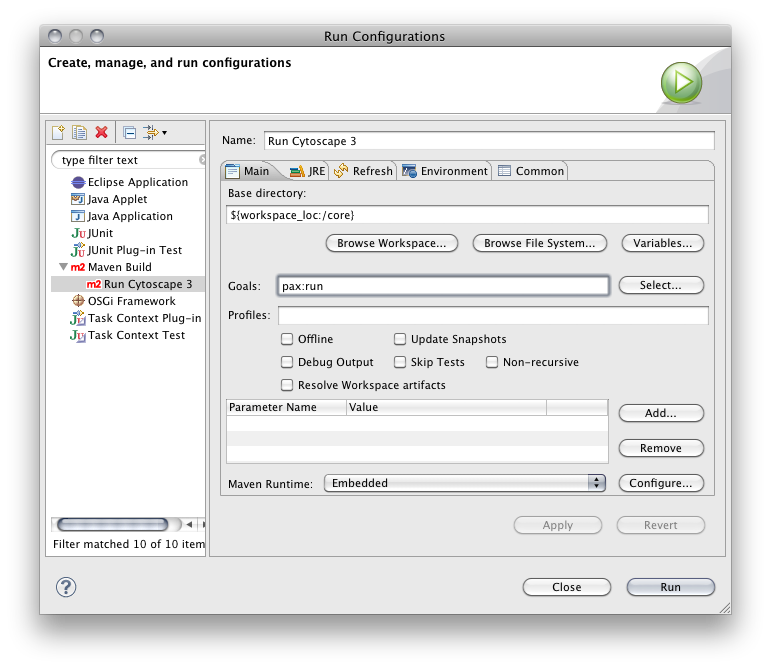
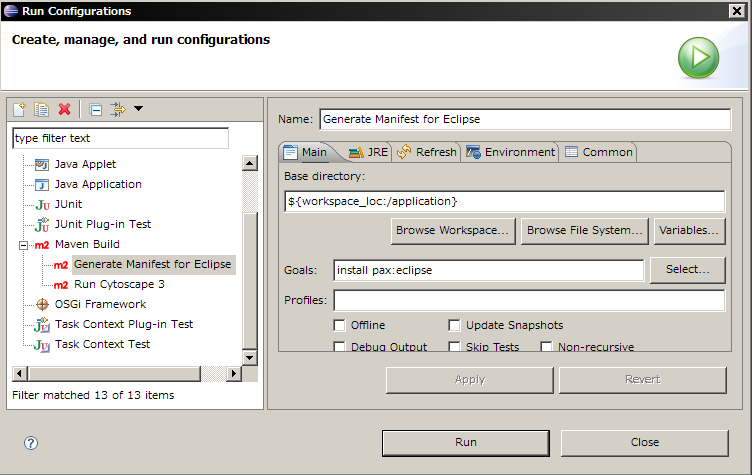
Select Run > Run Configurations...
Create new Maven Build
Name it Add OSGi Project Nature
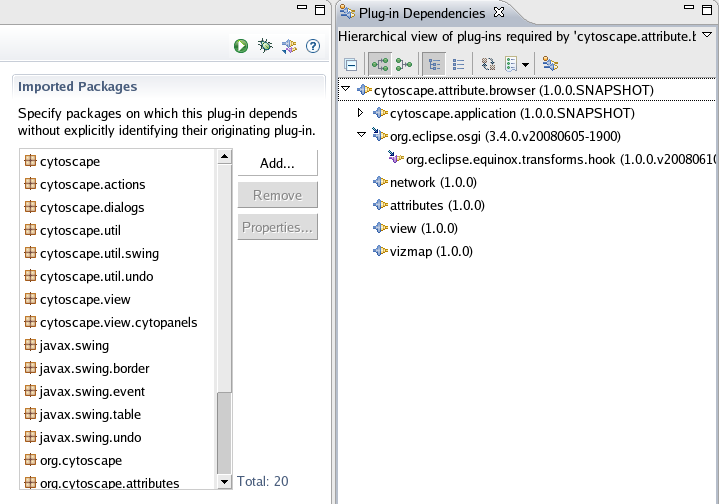
Click Browse Workspace and select project you want to add OSGi project nature. In this example, attribute browser project is selected.
Set pax:eclipse for goals
Apply and Run. This process may take 1-5 minutes depends on your network connection because it downloads source code of the dependent libraries from public repository.
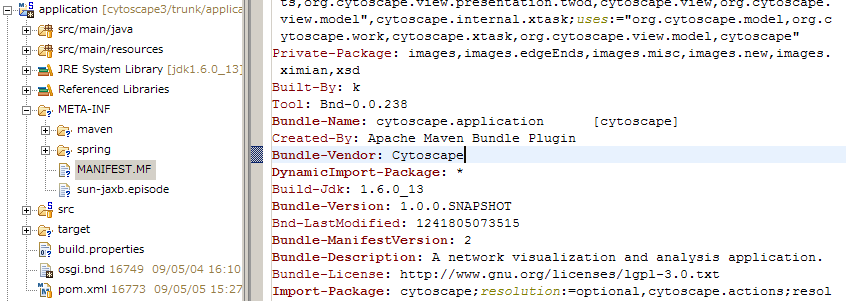
Once it is done, refresh the workspace (press F5). You can see META-INF directory and under the folder, you can find MANIFEST.MF file. This is the metadata
Editing this metadata file by writing BND statements is beyond the scope of this tutorial and will be discussed later in Plugin Development section. However by using this feature, you can see what packages are available for the selected project (bundle), and what packages will be exported as public API.
Step 4. Debug Cytoscape 3 on Eclipse with PAX Cursor
In this section, you will learn how to debug Cytoscape 3 on Eclipse.
First, let's set a sample break point in CytoscapeInit class, which is the class to startup Cytoscape. It is in application/src/main/java/cytoscape/CytoscapeInit.java. You can set breakpoint anywhere in the public boolean init(CyInitParams params) method.
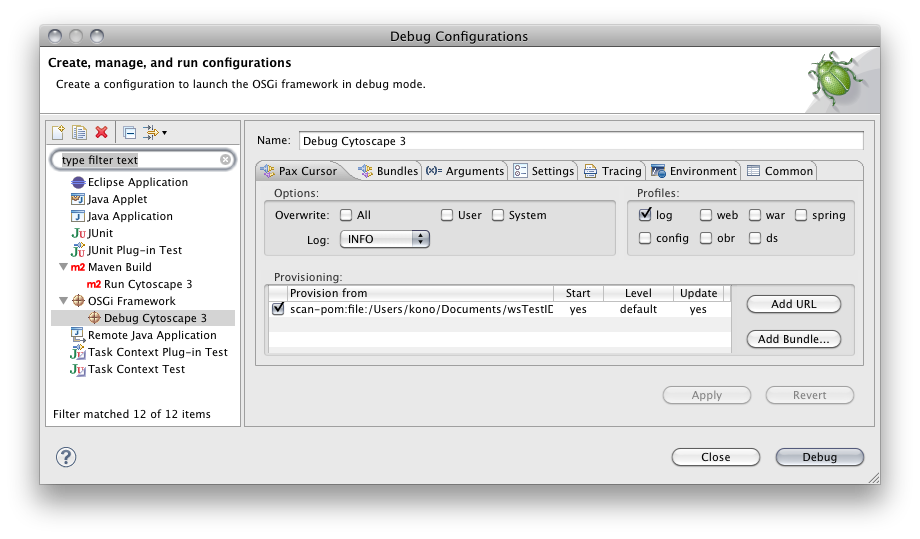
From Eclipse main window, select Run → Debug Configurations
Right click on OSGi Framework and select New
Name the configuration Cytoscape 3
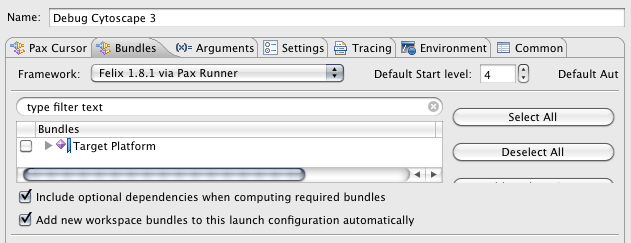
Switch framework to Felix 1.0.4
None of the files should be selected in the bundle list
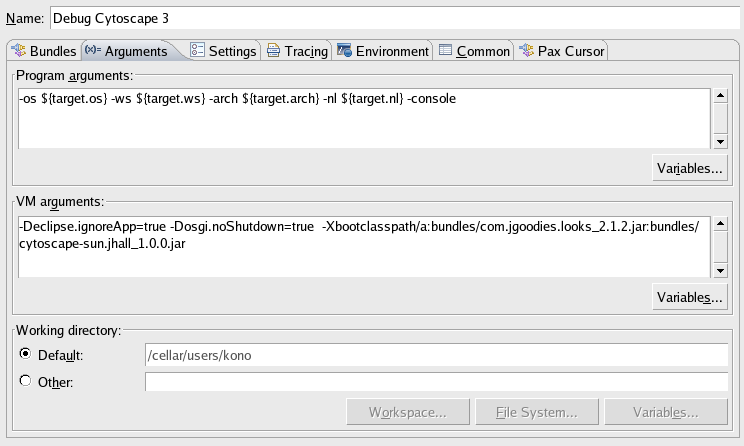
Click Arguments tab
Add the following VM Arguments
-Xbootclasspath/a:bundles/com.jgoodies.looks_2.1.2.jar:bundles/cytoscape-sun.jhall_1.0.0.jar
Click the Pax Cursor tab
Click Add Pom. Then select pom file under runner directory. To get the deploy-pom.xml file to show, you have to change the File of type ComboBox to have Any File.
Apply and Debug
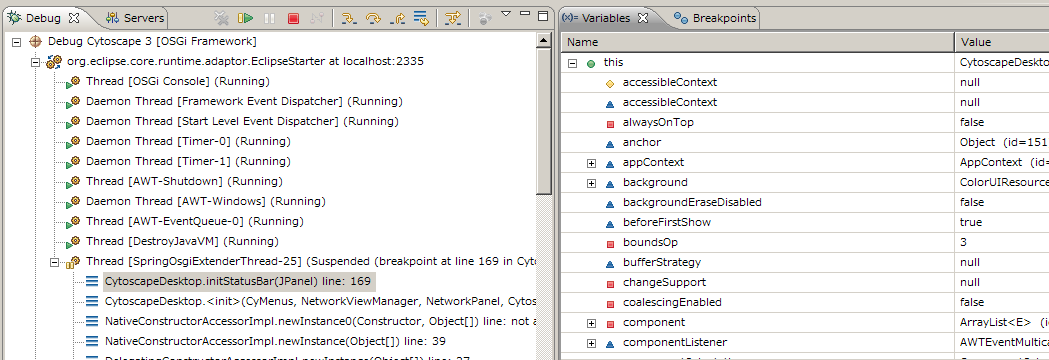
- Now you can see Cytoscape running on Felix and debugger shows current status of variables
Just like other applications, Eclipse debugger stops execution at the break-point you specified in the source code. To start again, you need to press green triangle (Resume button).
Basically, the debugging process is same as regular Java projects. Once you edit something in the source code, you can rebuild and deploy it to local repository by Maven install command. Then you can repeat the procedure above to check internal states of Cytoscape 3 by Eclipse debugger.
Working with Felix OSGi Console
Cytoscape 3 is an OSGi application and you can check the status of bundles from OSGi console. After you start Cytoscape 3 from Eclipse, you can see a prompt on the Console window (if not, hit enter in the console to show the prompt). The following is some useful command to check internal states of the OSGi system. For more information, please visit [Felix web site].
help - display help message
-> help bundlelevel <level> <id> ... | <id> - set or get bundle start level. cd [<base-URL>] - change or display base URL. headers [<id> ...] - display bundle header properties. help - display impl commands. install <URL> [<URL> ...] - install bundle(s). packages [<id> ...] - list exported packages. ps [-l | -s | -u] - list installed bundles. refresh [<id> ...] - refresh packages. resolve [<id> ...] - attempt to resolve the specified bundles. services [-u] [-a] [<id> ...] - list registered or used services. shutdown - shutdown framework. start <id> [<id> <URL> ...] - start bundle(s). startlevel [<level>] - get or set framework start level. stop <id> [<id> ...] - stop bundle(s). uninstall <id> [<id> ...] - uninstall bundle(s). update <id> [<URL>] - update bundle. version - display version of framework.
ps - List loaded bundles
-> ps START LEVEL 4 ID State Level Name [ 0] [Starting ] [ 0] System Bundle (1.0.4) [ 1] [Active ] [ 4] cytoscape.util.intr [cytoscape] (1.0.0.SNAPSHOT) [ 2] [Active ] [ 4] cytoscape.graph.fixed [cytoscape] (1.0.0.SNAPSHOT) [ 3] [Active ] [ 4] cytoscape.graph.dynamic [cytoscape] (1.0.0.SNAPSHOT) [ 4] [Active ] [ 4] org.cytoscape.network [org.cytoscape] (1.0.0.SNAPSHOT) [ 5] [Active ] [ 4] cytoscape.render.immed [cytoscape] (1.0.0.SNAPSHOT) [ 6] [Active ] [ 4] cytoscape.geom.spacial [cytoscape] (1.0.0.SNAPSHOT) [ 7] [Active ] [ 4] cytoscape.render.stateful [cytoscape] (1.0.0.SNAPSHOT) [ 8] [Active ] [ 4] cytoscape.geom.rtree [cytoscape] (1.0.0.SNAPSHOT) [ 9] [Active ] [ 4] cytoscape.render.export [cytoscape] (1.0.0.SNAPSHOT) [ 10] [Active ] [ 4] org.cytoscape.attributes (1.0.0.SNAPSHOT) [ 11] [Active ] [ 4] org.cytoscape.tunable (1.0.0.SNAPSHOT) [ 12] [Active ] [ 4] cytoscape.task [cytoscape] (1.0.0.SNAPSHOT) [ 13] [Active ] [ 4] phoebe.dnd [phoebe] (1.0.0.SNAPSHOT) [ 14] [Active ] [ 4] undo (1.0.0.SNAPSHOT) [ 15] [Active ] [ 4] org.cytoscape.view (1.0.0.SNAPSHOT) [ 16] [Active ] [ 4] org.apache.commons.cli [org.apache] (1.0.0.SNAPSHOT) [ 17] [Active ] [ 4] Apache Commons Lang (2.4.0) [ 18] [Active ] [ 4] org.cytoscape.layout (1.0.0.SNAPSHOT) [ 19] [Active ] [ 4] org.cytoscape.vizmap (1.0.0.SNAPSHOT) [ 20] [Starting ] [ 4] cytoscape.application [cytoscape] (1.0.0.SNAPSHOT) [ 21] [Resolved ] [ 4] JDOM DOM Processor (1.0.0) [ 22] [Resolved ] [ 4] Java Activation API (1.1.0) [ 23] [Installed ] [ 4] Concurrent (1.3.4) [ 24] [Resolved ] [ 4] Lowagie iText (2.0.8) [ 25] [Installed ] [ 4] csplugins.automatic.layout [csplugins] (1.0.0.SNAPSHOT) . . .
services - show list of available OSGi Services
services BUNDLE_ID - show detail of services abailable from the bundle
-> services 25 csplugins.automatic.layout [csplugins] (25) provides: ----------------------------------------------------- name = force-directed objectClass = org.cytoscape.layout.CyLayoutAlgorithm preferredMenu = Cytoscape Layouts service.id = 21 ---- name = circular objectClass = org.cytoscape.layout.CyLayoutAlgorithm preferredMenu = Cytoscape Layouts service.id = 22 ---- name = hierarchical objectClass = org.cytoscape.layout.CyLayoutAlgorithm preferredMenu = Cytoscape Layouts service.id = 23 ---- . . .
services -u - show usage of services.
-> services -u System Bundle (0) uses: ----------------------- org.osgi.service.startlevel.StartLevel org.cytoscape.layout (18) uses: ------------------------------- org.cytoscape.layout.CyLayoutAlgorithm org.cytoscape.layout.CyLayoutAlgorithm org.cytoscape.layout.CyLayoutAlgorithm org.cytoscape.layout.CyLayoutAlgorithm org.cytoscape.layout.CyLayoutAlgorithm
services -u BUNDLE_ID - show more detail about service usage of the bundle
org.cytoscape.layout (18) uses: ------------------------------- name = force-directed objectClass = org.cytoscape.layout.CyLayoutAlgorithm preferredMenu = Cytoscape Layouts service.id = 21 ---- name = circular objectClass = org.cytoscape.layout.CyLayoutAlgorithm preferredMenu = Cytoscape Layouts service.id = 22 ---- name = hierarchical objectClass = org.cytoscape.layout.CyLayoutAlgorithm preferredMenu = Cytoscape Layouts service.id = 23 ---- . . .
Note: The behaviour of Cytoscpae 3 running on Felix will be different from time to time. This is because budle loading timing is not properly controlled now. This will be solved by organizing bundle/service dependency. I'm working on this issue now and hopefully it will be solved soon.
Notes
Now you are ready to hack Cytoscape 3 on Eclipse. As you can see on your workspace, Cytoscape 3 is a collection of OSGi bundles. Each bundle is built as a Maven project and has a pom.xml file.
Currently, the desktop version of Cytoscape 3 is called application bundle. Under src/main/java you can see the source tree. You can edit these source files just like 2.x series, and create the bundle by running maven commands. To run them, you need to select (right click) the project name and then select maven command from Run As menu:
- maven clean - clean all classes from target directory.
- maven install - build everything under the selected project, create bundles, run tests, and install them to your local Maven repository
To run your modified code, you just need to do maven install and run debugger.
Troubleshooting
Somehow Cytoscape 3 Eclipse project is broken and I cannot compile it anymore
From terminal, run mvn pax:clean pax:eclipse. This cleans up everything and gives you a fresh workspace.
pax:provision fails. What should I do?
Cleaning up the working space (maven clean) and rebuilding it (maven install) solves this problem in most cases.
Cannot compile application bundle. Eclipse display some warning messages and cannot complete install goal
Comment out following lines in pom.xml in core directory:
<showWarnings>true</showWarnings> <showDeprecation>true</showDeprecation>
Eclipse and Cytoscape are frozen after splashscreen is displayed. What happened?
- On Windows, somehow Felix shell waits user's key entry after Cytoscape displays splashscreen. To proceed, you need to hit enter key in the console window (see below):
- On Windows, somehow Felix shell waits user's key entry after Cytoscape displays splashscreen. To proceed, you need to hit enter key in the console window (see below):
Appendix
Install Required Eclipse Plugins One by One
Subversion Client (Subclipse)
Cytoscape 3 source code is managed by Subversion. To access the repository, you need to install subversion client for Eclipse. In this tutorial, I'll use Subclipse. You can install the plugin from the eclipse's update manager. Do not forget to install SVNKit. Otherwise, you will get an error message when you try to check out repository ("Unable to load default SVN Client")
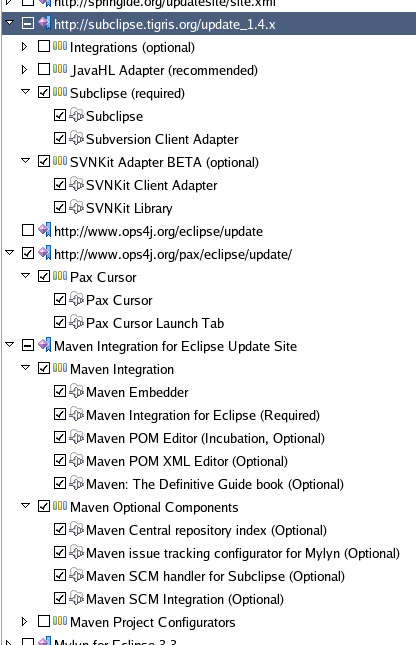
You can install the Subclipse plugin from Help > Software Updates... Click Available Software tab and add this URL by pressing Add Site button: http://subclipse.tigris.org/update_1.4.x
PAX Cursor
PAX Cursor is an Eclipse plugin developed by OPS4J developers. Basically, it is an Eclipse integration for PAX-Runner. By this plugin, you can run set of bundles on multiple platforms including Felix, Equinox, and Knopflerfish.
To install this Eclipse plugin, just follow the standard plugin installation procedure discussed above.
PAX Cursor Eclipse Update Site - http://www.ops4j.org/pax/eclipse/update/
Maven Integration for Eclipse (m2eclipse)
Cytoscape build system has been moved to Maven. To use maven from Eclipse, you need to follow this instruction. Even if you run all maven commands from Eclipse, you still need to install command-line version of Maven since Eclipse embedded version of maven have issues to execute some maven goals.
m2eclipse plugin update site - http://m2eclipse.sonatype.org/update/
This document is written by KeiichiroOno
If you have any questions, please e-mail me (kono at ucsd dot edu. English or Japanese)