<<TableOfContents: execution failed [Argument "maxdepth" must be an integer value, not "[1]"] (see also the log)>>
About this Document
This is an informal report on an investigation into possible technology sharing between Cytoscape and the InfoVis toolkit.
Status
![]() This document still under construction, but open for comments, as well.
This document still under construction, but open for comments, as well.
Background
The InfoVis Toolkit is an interactive graphics toolkit written by Jean-Daniel Fekete at INRIA (The French National Institute for Computer Science and Control). The toolkit implements nine (9) types of visualization: Scatter Plots, Time Series, Parallel Coordinates and Matrices for tables; Node-Link diagrams, Icicle trees and Treemaps for trees; Adjacency Matrices and Node-Link diagrams for graphs. Node-Link visualizations provides several variants (8 for graphs and 4 for trees). There are also a number of interactive controls and information displays, including dynamic query sliders, fisheye lenses, and excentric labels. Information about the InfoVis toolkit can be found at http://ivtk.sourceforge.net/
For the past several weeks, a number of us -- Melissa Cline, Scooter Morris, Alex Pico, Benno Schwikowski and Allan Kuchinsky -- have been corresponding with Jean-Daniel about the potential for Cytoscape to use components of InfoVis and vice versa. In looking at the InfoVis toolkit, we were impressed with the richness and sophistication of the repertoire of visualizations provided. A number of the most successful information visualization concepts and tools have been encapsulated in the toolkit in a very accessible way. There appears to be a basis for technology exchange between Cytoscape and InfoVis which would increase the power and usability of both software systems.
More recently, with Jean-Daniel's help, Allan has been doing a couple of prototyping experiments, in order to explore the feasibility of the toolkits interoperating and the compatibility of their respective object models.
To date, Allan has done two rough prototypes, which perform the following functions:
visualize a Cytoscape graph as an Adjacency Matrix in InfoVis. This involves converting a CyNetwork into an InfoVis graph structure and running the InfoVis tookit as a Cytoscape plugin.
implement Excentric Labels on top of a CyNetwork. Excentric labels are dynamically generated labels that cover the neighborhood around a node of interest. This is useful for navigating through dense networks.
The figure that follows(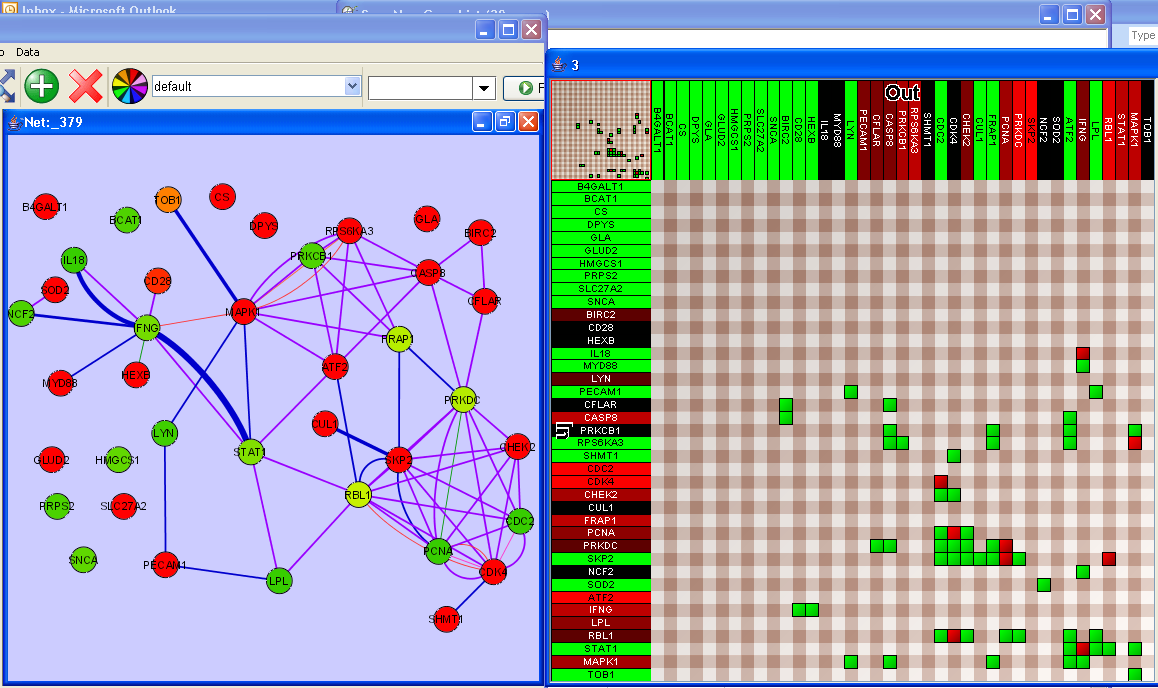 ) shows a Cytoscape Network next to an Adjacency Matrix view of the same network. In the matrix view on the right, the column labels are colored by out-degree of the node and the row labels are colored by the in-degree of the node, on a green-to-red scale. The cells in the matrix view are colored by the number of connections between the x,y pair of nodes corresponding to that cell.
) shows a Cytoscape Network next to an Adjacency Matrix view of the same network. In the matrix view on the right, the column labels are colored by out-degree of the node and the row labels are colored by the in-degree of the node, on a green-to-red scale. The cells in the matrix view are colored by the number of connections between the x,y pair of nodes corresponding to that cell.
The attached movie file (test_excentric.avi) runs a brief screen capture movie that shows the usage of excentric labels.
Discussion
It was relatively straightforward to achieve interoperability between Cytoscape and InfoVis. The initial matrix view prototype was easy to implement, the biggest challenge being getting the right jar structure. Jean-Daniel was able to accomplish this pretty quickly.
There are three issues with the current implementation of the excentric labels prototype.
It has been difficult to get the endpoints of the lines emanating from the excentric labels to coincide exactly with the centerpoints of CyNodes that they correspond to. This is due to limitations of the Cytoscape renderer. There are methods for mapping a point in CytoscapeDesktop coordinates to the corresponding position in CyNetwork coordinates, but no inverse methods for mapping a point in CyNetwork coordinates back to CytoscapeDesktop. Allan has attempted to build such methods, without success. One source of difficulty is that the mapping of desktop to network coordinates is destructive of the original transforms.
- There is a flickering effect when the density of the labels in an area is over a certain amount. It's possible that the labels are overwriting each other on the display.
- Cytoscape slows appreciably when the excentric labels are activated. This may likely be due to faulty logic in the prototype code itself, possibly the unintentional introduction of tight loops. Some profiling is needed to better characterize the problem.
It should be noted that the InfoVis toolkit is currently a proof-of-concept prototype. A more robust version is planned, with work beginning at INRIA in Fall of 2006. One intention of this investigation is to provide feedback that can influence the direction of the next version of InfoVis.
It should also be noted that Jean-Daniel has been extremely helpful and appears to be quite motivated to work with us.
The Plan Going Forward
Allan, Melissa, Benno, and Jean-Daniel will meet in Paris during the first week of September to review progress and discuss next steps. We are thinking of grounding any collaboration in a small set of applications. One of the goals of the Paris meetings is to flesh out a set of candidate applications which could be presented at the Cytoscape Developers Retreat in mid-October. The ideal would be to get to the point that people are ready to further explore concrete interactions after the fall meeting.
The following subsections discuss candidate applications. ![]() This section still under construction, but open for comments, as well. Additional application ideas would be very helpful.
This section still under construction, but open for comments, as well. Additional application ideas would be very helpful.
Candidate Application: Post-translational Modifications
- use Cytoscape to display similarity graphs colored by PTMs
- different colorings for different types of PTMs
- work into subcellular localization
use InfoVis for linked adjacency matrix view(s) with dimensions of
- Blast sequence similarity
- RMS (ruby squared 3D structure)
- protein function
Note: this similar problem to representing spice variants in Cytoscape, i.e. a subset of the subgene concept that GenMAPP/Agilent are implementing. So this is a natural offshoot of the current GenMAPP/Agilent project, perhaps a followon
Candidate Application: InfoVis Dynamic Queries as a Filtering interface
Implement a DynamicQuery interface, a la SpotFire, InfoVis toolkit, as an alternative filtering capability. Use a set of RangeSlider widgets, one per desired attribute, with filtering performed by an ANDing of the individual attribute/slider queries. Have filtering result in hiding/showing of nodes, rather than selection of nodes. The InfoVis toolkit has classes for Dynamic Queries and range sliders, that we can utilize.
Comments
AllanKuchinsky, 8/01/2006 - This is a sample comment to get things started