LinkOut PlugIn
The LinkOut plugin provides a mechanism to link nodes to external web resources within the Cytoscape viewer. Right-clicking on a node opens a popup menu with a list of web links. Selecting a link initiates a search at the specified website using the node label as the query term.
LinkOut includes a number of predefined links to commonly used web resources such as NCBI and UniProt as well as species-specific databases. In addition to the default links, users can customize the LinkOut menu by adding (or removing) links without the need to restart Cytoscape.
If you don't have the LinkOut plugin with your version of Cytoscape, you can download is from here: <TODO: attach jar>. LinkOut currently only works with Cytoscape version 2.3 or later.
LinkOut properties format
Similar to other properties LinkOut links are listed as ‘key’-‘value’ pairs where key specifies the name of the link and value is the URL link. The LinkOut menus are organized in a hierarchical structure that is specified in the key. All key terms start with the keyword linkouturl.
For example, the following entry: linkouturl.yeast.SGD=http://db.yeastgenome.org/cgi-bin/locus.pl?locus\=%ID%
places the SGD link under the yeast submenu. This link will appear in Cytoscape as:
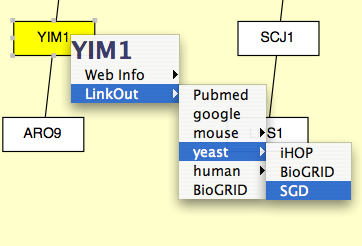
In a similar fashion one can add new links under existing submenu or place them in a new submenu.
The %ID% string in the URL is a place-holder for the node label. When the menu is generated this marker is substituted with the node label. In the above example the generated SGD link for YIM protein is: linkouturl.yeast.SGD=http://db.yeastgenome.org/cgi-bin/locus.pl?locus\=YIM1
Currently there is no mechanism to check whether the constructed URL query is correct and if the node label is meaningful. Similarly there is no ID mapping between various database identifiers. For example, a link to NCBI Entrez from a network that uses ensembl gene identifiers as node label will produce a link to Entrez using ensembl ID that will return an empty page. It is the users responsibility to ensure that the node label that is used as the search term in the URL link will result in a meaningful link. Likewise, most links will not work for all network since they use different identifiers. Links can be removed using the Preference Dialog.
Adding new links
Similar to other properties LinkOut links can be modified using the Cytoscape Preferences Dialog (access from the Edit->Preferences menu). To add a links open the Preferences Dialog, in the Properties window click the ‘Add’ button. In the Property name box add the name of the link. All LinkOut link names begin with the linkouturl prefix followed by the submenus path ending with the name of the link. For example, to add a Reactome link to Biological resources submenu enter the property name as: linkouturl.Biological resources.Reactome.
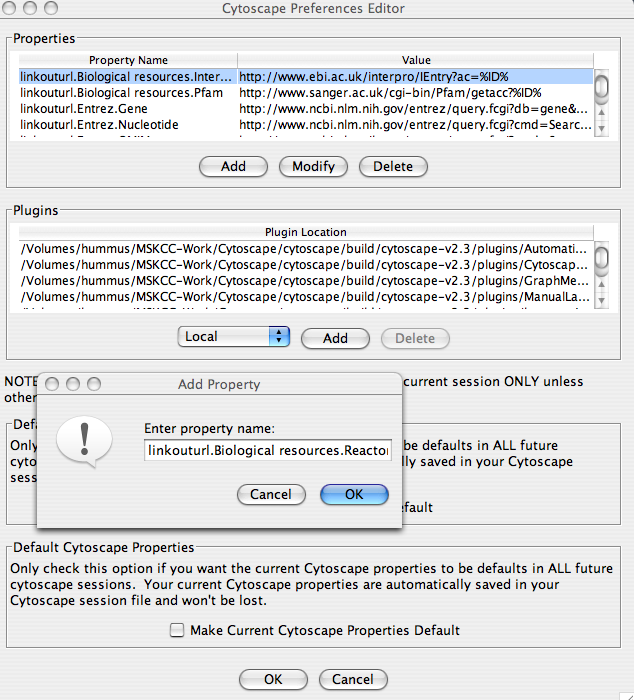
It is also possible to create a new submenu(s) e.g. linkouturl.Pathway DB.Reactome. In the property value window enter the URL link. Remember that the query value is replaced with %ID%, which will be replaced with the node label.
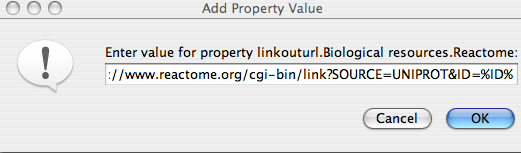
To save the modified links for future sessions select the 'Make Current Cytoscape Properties Default' option.