Web Service Client Manager
Cytoscape 2.6.0 has a new feature called Web Service Client Manager. This is a framework to manage various kinds of web service clients in Cytoscape. By using web service clients, users can access remote data sources easily.
What is a Web Service?
A web service is a standardized, platform-independent mechanism for machines to interact over the network. These days, many major biological databases publish their data with web service API:
List of Biological Web Services: http://taverna.sourceforge.net/services
Web Services at the EBI: http://www.ebi.ac.uk/Tools/webservices/
This enables developers to write a program to access these services. Cytoscape core developer team have developed several sample web service clients using this framework. Cytoscape supports many web services including:
PSICQUIC: Standard web service for biological interaction data sets. As of July 2010, the following data providers support PSICQUIC:
Pathway Commons: an open source portal, providing access to multiple integrated data sets, including: Reactome, IntAct, HPRD, HumanCyc, MINT, the MSKCC Cancer Cell Map, and the NCI/Nature Pathway Interaction database.
NCBI Entrez Gene: a public database of genes, including annotation, sequence and interactions.
Biomart: an open source biological database engine. Useful for ID/Name mapping.
All of these clients are available as Plugins and users can install them through Plugin Manager.
In the following sections, users learn how to import network from extrenal databases.
Getting Started
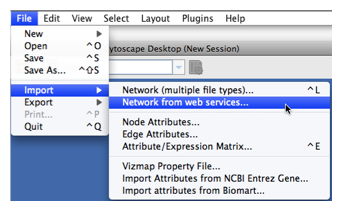
To get started, select: File → Import → Network from web services...
Tip: View the animation demo for importing networks from web services. |
Example #1: Retrieving Protein-Protein Interaction Networks from IntAct
Select: File → Import → Network from web services...
From the pull-down menu, select the IntAct Web Service Client.
- Enter one or more search terms, such as BRCA1
- Click the Search button.
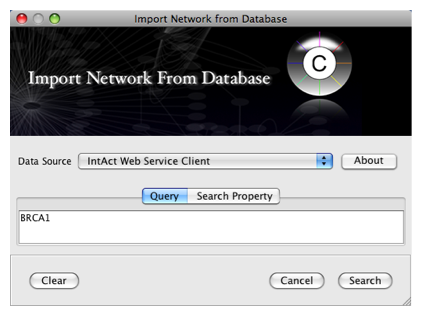
After confirming the download of interaction data, the network of BRCA1 will be imported and visualized.
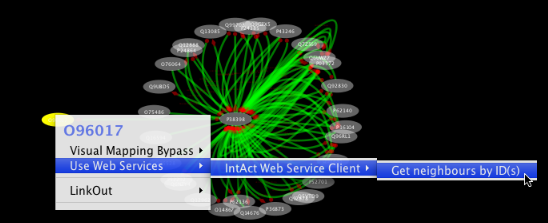
Tip: Expanding the Network: Several of the Cytoscape web services provide additional options in the node context menu. To access these options, right-click on a node and select "Use Web Services." For example, in the screenshot to the right, we have loaded the BRCA1 network from IntAct, and have chosen to merge this node's neighbors into the existing network.
Example #2: Retrieving Protein-Protein Interaction Networks from NCBI Entrez Gene
An entry of NCBI Entrez Gene has a section called Interactions. NCBI web service client uses this section to build networks.
Select: File → Import → Network from web services...
From the pull-down menu, select the NCBI Web Service Client.
Enter free-keywords. For example, type human muscular dystrophy.
- Click the Search button.
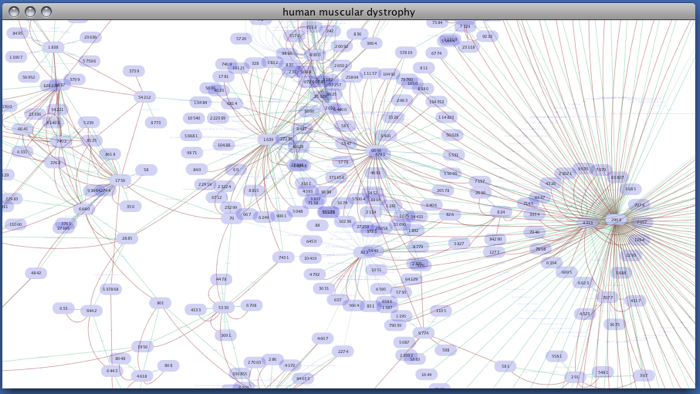
Network generated from Entrez Gene data: The network above is generated from interaction data matching the keyword human muscular dystrophy. Edge color represents data source type (BIND, BioGRID, or HPRD).
Note: since NCBI client extracts interaction data from a huge dataset, it takes a long time (30 seconds - 5 minutes, depends on machine specifications and network connection) to import large set of interactions.
Example #3: Retrieving Pathways and Networks from Pathway Commons
Select: File → Import → Network from web services...
- From the pull-down menu, select the Pathway Commons Web Service Client.
Then, follow the three-step process outlined below:
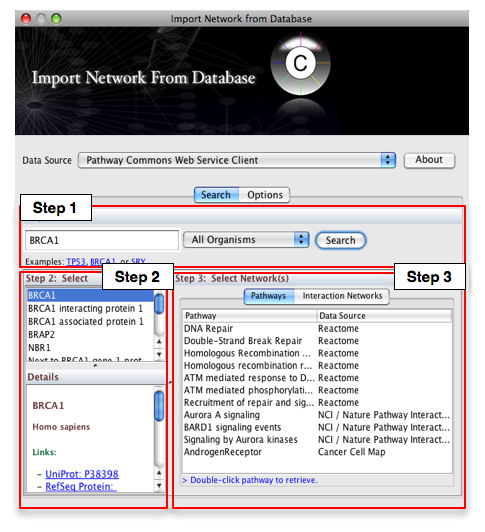
- Step 1: Enter your search term; for example: BRCA1
- Step 2: Select the protein or small molecule of interest. Full details regarding each molecule is shown in the bottom left panel.
- Step 3: Download a specific pathway or interaction network.
Downloading Pathways and Interaction Networks
In Step 3, you can simply double-click on a pathway of interest, or click on the Interaction Networks tab. The Interaction Networks tab enables you to filter interactions by data source and/or interaction type. For example, you can choose to restrict your network to direct physical interactions from HPRD and MINT only:
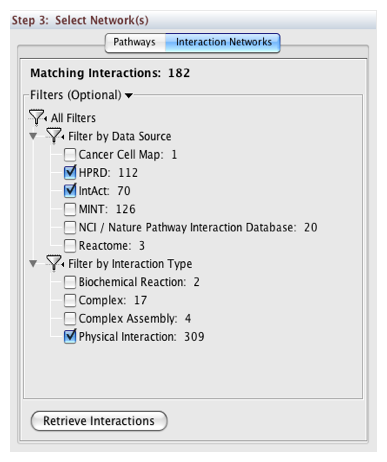
Pathway Commons Options
You can configure access options from the Options tab. There are two retrieval options:
- Simplified Binary Model: Retrieve a simplified binary network, as inferred from the original BioPAX representation. In this representation, nodes within a network refer to physical entities only, and edges refer to inferred interactions.
- Full Model: Retrieve the full model, as stored in the original BioPAX representation. In this representation, nodes within a network can refer to physical entities and interactions.
By default, the simplified binary model is selected.
Future Directions
As additional web service clients become available, they will be made available via the Cytoscape Plugin Manager. Once installed, these web service clients will be centrally accessible via the same steps defined above:
File → Import → Network from web services...
Import Attributes from External Database
Some of the web service clients can import attributes from external databases. BioMart client is an example. You can install it from Plugin Manager.
Example 1: Import Additional ID Sets and Annotations from BioMart
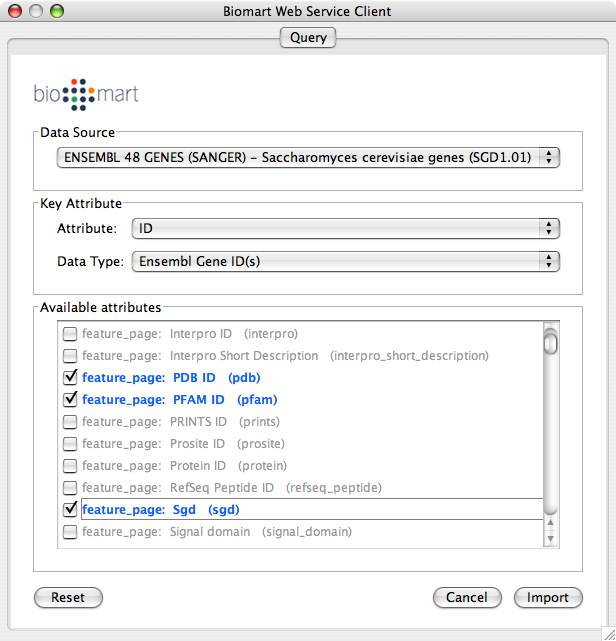
Load a network. In this example, we use galFiltered.sif in sampleData directory.
File → Import → Import Attributes from BioMart...
Select Data Source. Since galFiltered.sif is a yeast network, select yeast dataset.
For Key Attribute section, select ID for Attribute and Data Type should be Ensembl Gene ID. Attribute is the list of available attributes in current Cytoscape session and Data Type is the type of ID set of the attribute. In this case, Cytoscape uses ID as the key for mapping. Because the sample network galFiletred.sif uses Ensemble Gene ID for its node ID, like YOR072W, you need to select Ensembl Gene ID for Data Type. So you need to know the type of ID set (Entrez Gene ID, UniProt Unified Acc. Number, Ensemble Gene ID, etc.) of the attribute selected in the Attribute box.
Select attributes you want to import. (Note: You cannot select too many attributes at once because BioMart server has maximum number of selectable annotations.)
Press Import.
Now you can see the newly imported attributes on the Attribute Browser. You may see some attribute names ends with -TOP if there are multiple attribute values for a node. This is an attribute taken from the first entry of the original list attribute.
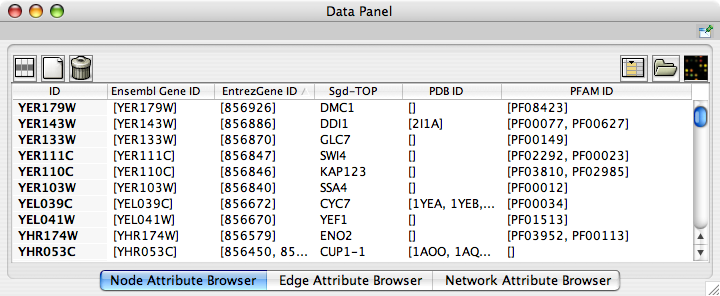
Example 2: Import Annotations from NCBI Entrez Gene Database
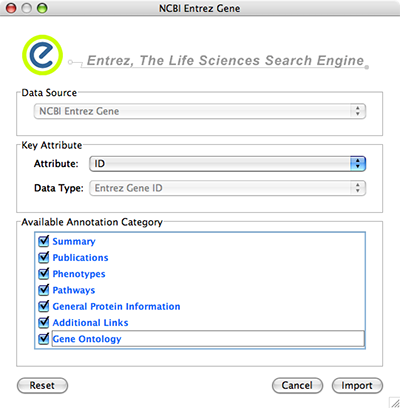
NCBI Entrez Gene database () can be used as network data source and annotation repository. You can use NCBI web service client to import gene annotations from Entrez Gene.
File → Import → Import attributes from NCBI Entrez Gene...
Data Source is fixed to NCBI Entrez Gene.
Data Type is also fixed to Entrez Gene ID. This means Cytoscape attribute selected in Attribute list should be Entrez Gene ID.
Select Annotation Category. If you select a category, all of the annotation under the category will be imported (i.e., multiple Cytoscape attributes will be created for each category).
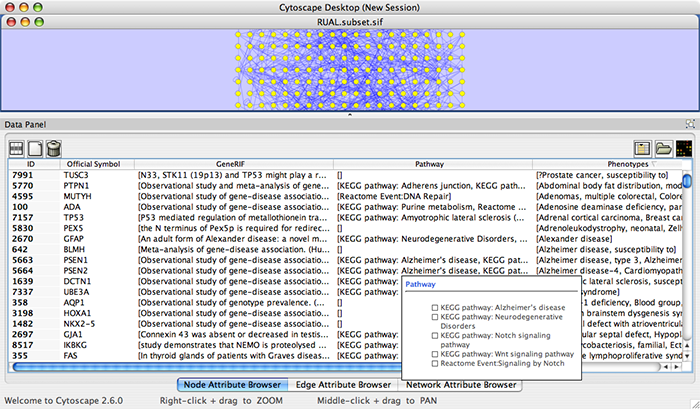
Press Import. The following is a sample human network (RUAL.subset.sif) annotated by NCBI client.
Use Multiple Services in a Workflow
Web services are useful when you combine the result from multiple data sources.
Example: Import and Annotate Networks
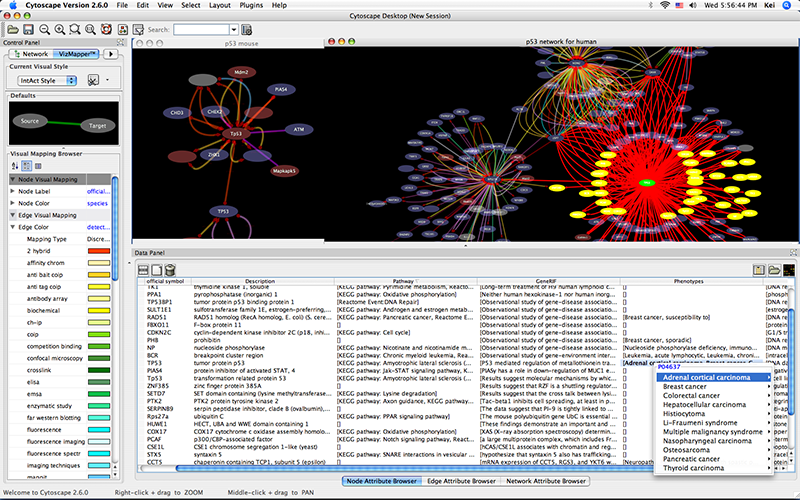
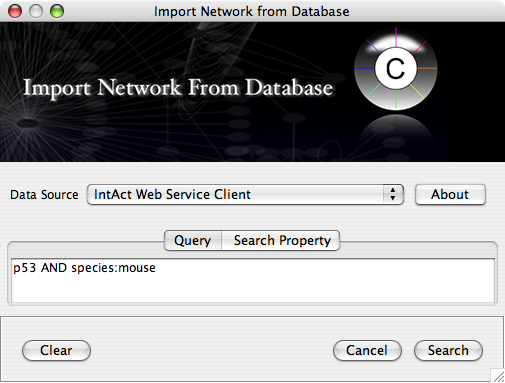
Import network from IntAct using keyword. In this example, type p53 AND species:mouse.
Import human orthologs from BioMart.
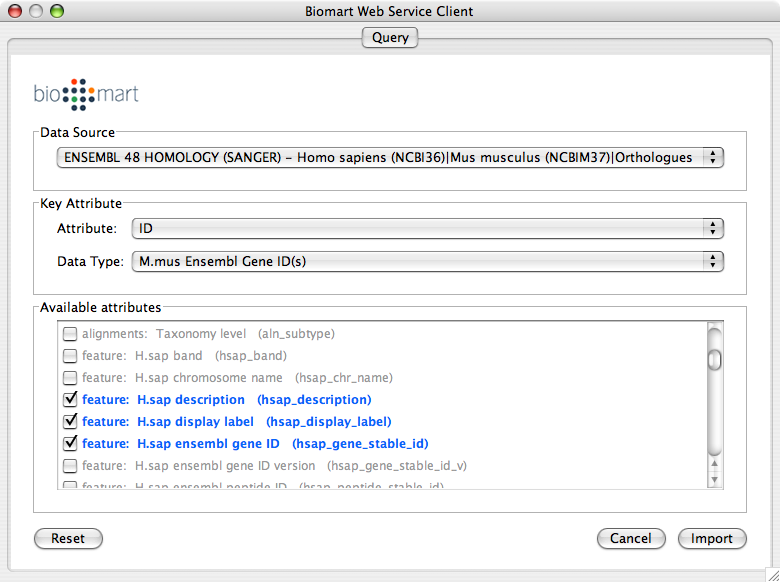
Show the othologs as the list of Ensembl Gene ID on the Data Panel. Copy them and use them as the query for IntAct.
Import Entrez Gene ID from BioMart. Use ensembl attribute for the mapping key.
- Import annotations from NCBI. The resulting networks looks like the following: