Scripting Feature is Deprecated!
Due to limitations of JVM languages and OSGi, this feature is deprecated. Please use cyREST instead.
Scripting in Cytoscape 3
Scripting language support is an experimental feature in Cytoscape 3. Java runtime environment comes with built-in scripting language support and Cytoscape uses it to execute tasks written in standard languages such as JavaScript.
This feature is for advanced users. If you just want to automate some basic tasks, such as loading network, you can use Command instead.
Run Cytoscape from Command Line
To use Scripting feature, you need to start Cytoscape from console. In Cytoscape application directory, you can find cytoscape.sh (for Windows, cytoscape.bat). This is a shell script to start Cytoscape from command line.
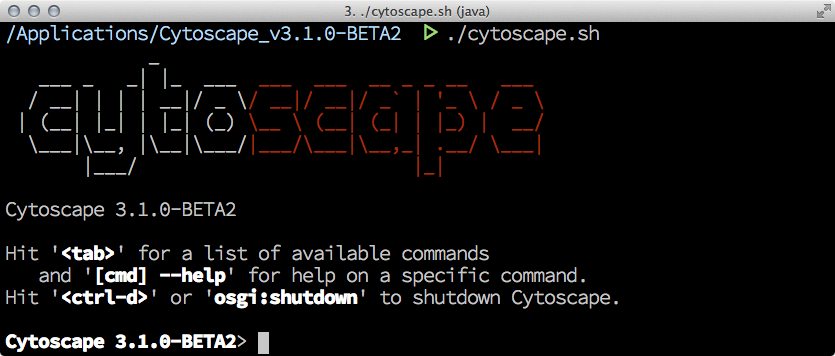
Once you run Cytoscape from command line, you can use Apache GOGO OSGi Shell. If you type:
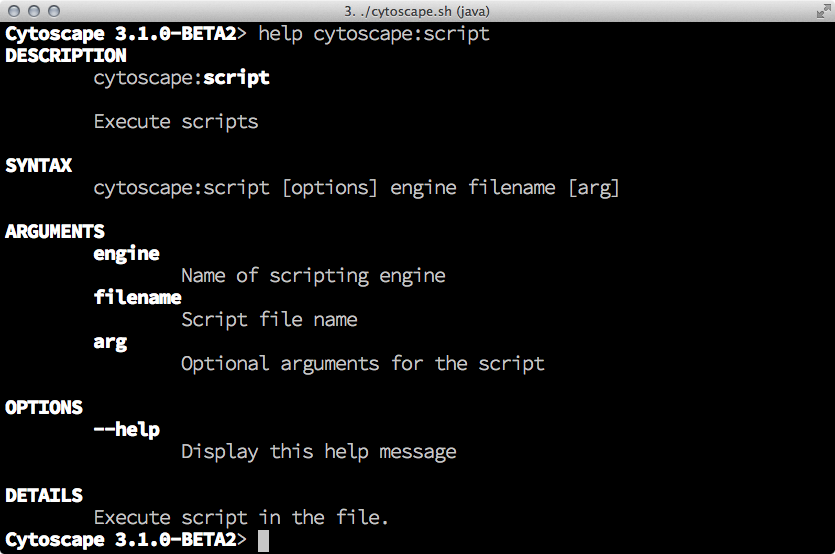
help cytoscape:script
the shell displays help message how to use script files in Cytoscape.
JavaScript
JavaScript is supported by default. To run your script, type:
cytoscape:script javascript SCRIPT_FILE_NAME
also, you can pass optional command line arguments.
Sample Script: Create complete graph
// Generate a complete graph
// Extract command-line arguments
var numberOfNodes = args[0];
if(numberOfNodes == null)
numberOfNodes = 10;
// Create new network
var newNetwork = cyAppAdapter.getCyNetworkFactory().createNetwork();
newNetwork.getRow(newNetwork).set("name", "Complete Graph with " + numberOfNodes + " Nodes");
// Register the new network to Network Manager
cyAppAdapter.getCyNetworkManager().addNetwork(newNetwork);
// Add nodes
var nodes = [];
for( i = 0; i < numberOfNodes; i++) {
var nodeName = "Node " + i;
var node = newNetwork.addNode();
newNetwork.getRow(node).set("name", nodeName);
nodes[i] = node;
}
// Add edges
var edgeCount = 0;
for( i = 0; i < numberOfNodes; i++) {
var source = nodes[i];
for( j = 0; j < numberOfNodes; j++) {
var target = nodes[j];
if(newNetwork.containsEdge(source, target) == false &&
newNetwork.containsEdge(target, source) == false && j != i) {
var edge = newNetwork.addEdge(source, target, true);
newNetwork.getRow(edge).set("name", "Edge " + edgeCount++);
newNetwork.getRow(edge).set("interaction", "interacts_with");
}
}
}This script is available here: https://gist.github.com/keiono/7915968
By default, cyAppAdapter object will be injected, just like Simple Apps. You can access basic functions from this object. For more information, please read Cytoscape API document.
Experimental: Adding Other JSR 223 Compatible Scripting Engines
Warning: This method directly adds ALL classes provided by the library to your classpath. Maybe there are some side-effects to other OSGi Apps.
In theory, you can use full features of scripting languages written on top of JVM by adding JSR 223 (Scripting for the Java Platform) compatible Scripting Engines to Cytoscape runtime. The description below is how to add new scripting languages to Cytoscape 3.
Adding New Scripting Languages to Cytoscape
Scripting feature in Cytoscape is very simple. Cytoscape just searches available JSR 223 compatible Scripting Engines and use specified Engine when user runs a script file. To add a new Scripting Engine, simply add the JAR files to CYTOSCAPE_APPLICATION_DIR/framework/lib/ext/. For example, if you want to add Python (Jython) scripting engine, copy the standalone jar file to this directory:
Now it's ready to use. Just specify scripting engine in the command line arguments.
Python
Support for Python can be added by importing Jython Scripting Engine. Simply copy standalone version of JAR file to the ext directory. Access to cyAppAdapter is also available for Python.
Sample Script
This script creates a complete graph with 20 nodes:
1 #
2 # Python Script to generate complete graph
3 #
4
5 NUM_NODES = 20
6
7 print "Creating complete graph with " + str(NUM_NODES) + " nodes..."
8
9 new_network = cyAppAdapter.getCyNetworkFactory().createNetwork()
10 new_network.getRow(new_network).set("name", "Complete Graph Created by Python Script")
11 cyAppAdapter.getCyNetworkManager().addNetwork(new_network);
12
13 # Add nodes
14 nodes = [];
15 for i in range (NUM_NODES):
16 node_name = "Node " + str(i)
17 node = new_network.addNode()
18 new_network.getRow(node).set("name", node_name)
19 nodes.append(node)
20
21 # Add edges
22 edge_count = 0;
23 for source in nodes:
24 for target in nodes:
25 if new_network.containsEdge(source, target) is False \
26 and new_network.containsEdge(target, source) is False \
27 and source is not target:
28 edge = new_network.addEdge(source, target, True)
29 edge_count = edge_count + 1
30 new_network.getRow(edge).set("name", "Edge " + str(edge_count))
31 new_network.getRow(edge).set("interaction", "interacts_with")
32
To run, type:
cytoscape:script python /ABS_PATH_TO_SCRIPT/complete_graph.py
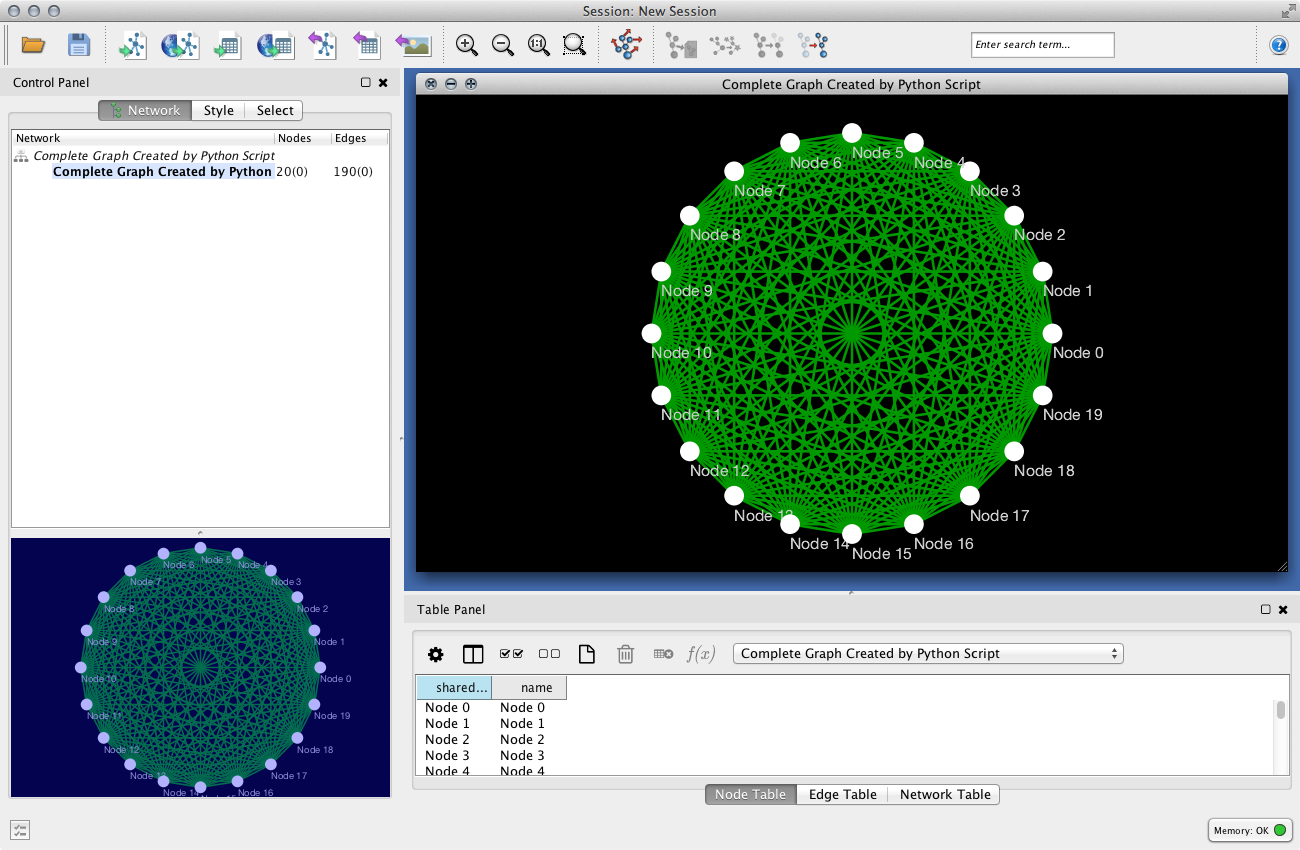
Source code is available here: https://gist.github.com/keiono/7919559
Clojure
The original Clojure distribution does not contain JSR 223 implementation. You need to build this library to run Clojure scripts.
; Node Names - sequence of integers.
(def nodenames (take 500 (range)))
; Create new network and register it to manager
(def newnet(. (. cyAppAdapter getCyNetworkFactory) createNetwork))
(.. newnet (getRow newnet) (set "name" "Random Network Created by Clojure Script"))
(.. cyAppAdapter (getCyNetworkManager) (addNetwork newnet))
; Create nodes
(doseq [nodename nodenames]
(.. newnet (getRow (. newnet addNode))
(set "name" (str nodename))))
; Add random edges
(def nodes (. newnet getNodeList))
(doseq [node nodes]
(. newnet addEdge node (rand-nth nodes) false))https://gist.github.com/keiono/7938418
Tips for Debugging Your Scripts
Log
To display log messages, use this command:
tail -f ~/CytoscapeConfiguration/3/framework-cytoscape.log
Known Issues
Since dependencies are added in Classpath, you cannot access static methods in Cytoscape core API. The only way to access those is put actual API JAR files to system classpath.