|
Size: 3056
Comment:
|
Size: 3725
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 1: | Line 1: |
|
Describe Cytoscape_3/UserManual/Preferences here. |
|
| Line 6: | Line 4: |
| The Cytoscape Properties editor, accessed via Edit → Preferences → Properties…, is used to specify general and default properties. Any changes made to these properties will be saved in the file cytoscape3.props in the .cytoscape directory of user's home. Properties are now stored in Cytoscape session files, so changes to general properties will be saved as part of the current session. | The Cytoscape Properties editor, accessed via Edit → Preferences → Properties…, is used to specify general and default properties. Any changes made to these properties will be saved in the file {{{cytoscape3.props}}} in the {{{CytoscapeConfiguration}}} directory of user's home. Properties are now stored in Cytoscape session files, so changes to general properties will be saved as part of the current session. |
| Line 14: | Line 12: |
| == Managing Bookmarks == | |
| Line 15: | Line 14: |
|
=== Setting Default Properties === It is possible to alter the default properties for Cytoscape. Edit the properties via Edit → Preferences → Properties... and check the Make Current Cytoscape Properties Default checkbox. This will save the current properties to the {{{.cytoscape}}} directory, where they will then be applied to all of your Cytoscape sessions from that point on. Otherwise, Cytoscape will automatically save the properties used in a particular session inside its {{{.cys}}} session file, while the default properties will be applied at the beginning of subsequent sessions. == Managing Bookmarks == |
|
| Line 24: | Line 17: |
| There are currently two types of bookmarks: network and annotation. Network bookmarks are URLs pointing to network files available on the Internet. These are nomal networks that can be loaded into Cytoscape. The annotation bookmarks are URLs pointing to ontology annotation files. The annotation bookmarks are only used when importing an ontology. | There are currently several types of bookmarks (based on data categories), including network and table. Network bookmarks are URLs pointing to network files available on the Internet. These are normal networks that can be loaded into Cytoscape. The table bookmarks are URLs pointing to attribute files. |
| Line 27: | Line 20: |
| You can define and configure a proxy server for Cytoscape by going to Edit → Preferences → Proxies… . | You can define and configure a proxy server for Cytoscape by going to Edit → Preferences → Proxy Settings… . |
| Line 30: | Line 23: |
| After the proxy server is set, all network traffic related to loading a network from URL will pass through the proxy server. Other plugins use this capability as well. The proxy settings are saved in {{{cytoscape.props}}}. Each time you click the Update button after making a change to the proxy settings, an attempt is made to connect to a well known site on the Internet (e.g., google.com) using your settings. For both success and failure you are notified and for failure you are given an opportunity to change your proxy settings. | After the proxy server is set, all network traffic related to loading a network from URL will pass through the proxy server. Other plugins use this capability as well. The proxy settings are saved in {{{cytoscape3.props}}}. Each time you click the OK button after making a change to the proxy settings, an attempt is made to connect to a well known site on the Internet (e.g., google.com) using your settings. For both success and failure you are notified and for failure you are given an opportunity to change your proxy settings. |
| Line 32: | Line 25: |
| If you no longer need to use a proxy to connect to the Internet, simply uncheck the Use Proxy checkbox and click the Update button. |
If you no longer need to use a proxy to connect to the Internet, simply set the Proxy type to 'direct' and click the OK button. == Managing Group view == The configuration of Cytoscape group view may also be edited through Edit → Preferences → Group preferences… {{attachment:groupViewSetting.png}} == Managing Rendering Engine Settings == Cytoscape's rendering engine uses different thresholds to determine how much detail to use when rendering a graph. When these thresholds are exceeded, the engine stops drawing some features on the graph. The following table summarizes the property names of these thresholds and how they affect rendering: || render.coarseDetailThreshold || When the number of visible nodes exceeds this value the rendering engine renders nodes and edges with minimal detail.|| || render.nodeBorderThreshold || When the number of visible nodes exceeds this value the rendering engine stops rendering node borders.|| || render.nodeLabelThreshold || When the number of visible nodes exceeds this value the rendering engine stops rendering node labels.|| || render.edgeArrowThreshold || When the number of visible edges exceeds this value the rendering engine stops rendering edge arrows.|| || render.edgeLabelThreshold || When the number of visible edges exceeds this value the rendering engine stops rendering edge labels.|| To force Cytoscape to use full detail always, enable the menu item View → Show Graphics Details. |
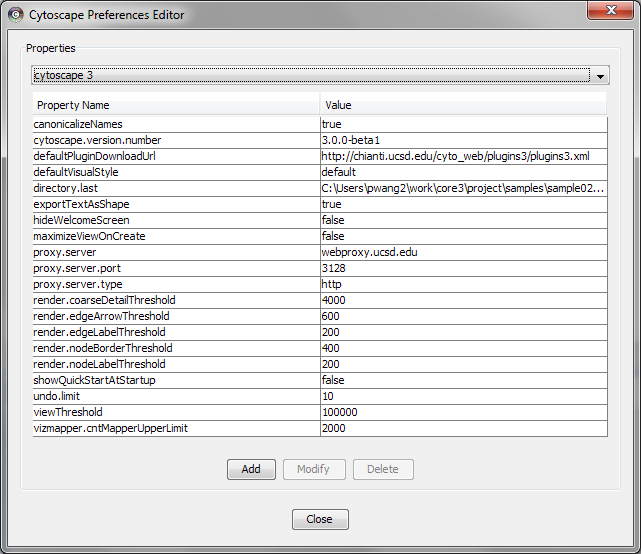
Managing Properties
The Cytoscape Properties editor, accessed via Edit → Preferences → Properties…, is used to specify general and default properties. Any changes made to these properties will be saved in the file cytoscape3.props in the CytoscapeConfiguration directory of user's home. Properties are now stored in Cytoscape session files, so changes to general properties will be saved as part of the current session.
Cytoscape properties are configurable via Add, Modify and Delete operations.
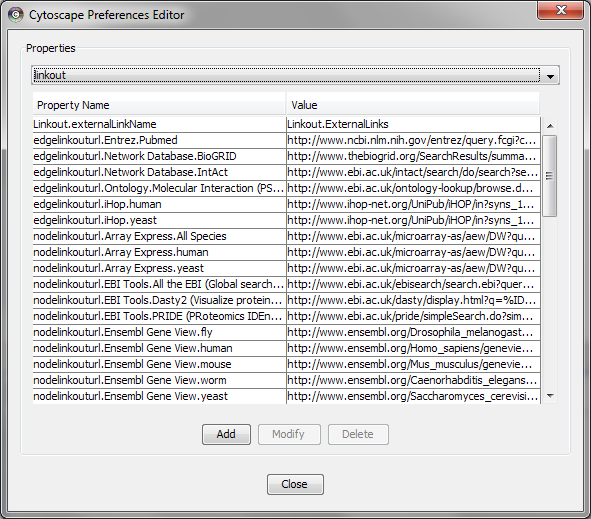
App properties may also be edited in the same way as editing Cytoscape properties. For example, to edit the properties of linkout, select 'linkout' from the combobox of the Preferences Editor.
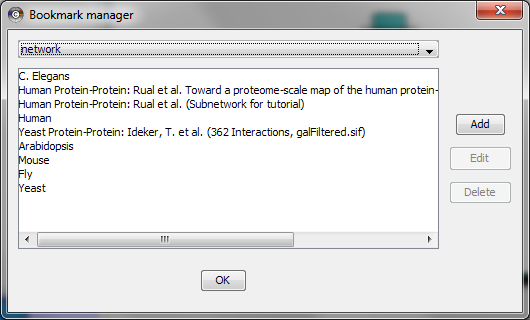
Managing Bookmarks
Cytoscape contains a pre-defined list of bookmarks, which point to sample network files located on the Cytoscape web server. Users may add, modify, and delete bookmarks through the Bookmark manager, accessed by going to Edit → Preferences → Bookmarks… .
There are currently several types of bookmarks (based on data categories), including network and table. Network bookmarks are URLs pointing to network files available on the Internet. These are normal networks that can be loaded into Cytoscape. The table bookmarks are URLs pointing to attribute files.
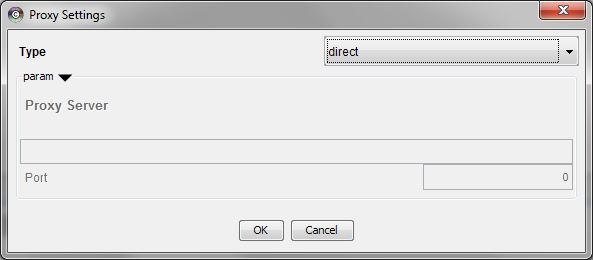
Managing Proxy Servers
You can define and configure a proxy server for Cytoscape by going to Edit → Preferences → Proxy Settings… .
After the proxy server is set, all network traffic related to loading a network from URL will pass through the proxy server. Other plugins use this capability as well. The proxy settings are saved in cytoscape3.props. Each time you click the OK button after making a change to the proxy settings, an attempt is made to connect to a well known site on the Internet (e.g., google.com) using your settings. For both success and failure you are notified and for failure you are given an opportunity to change your proxy settings.
If you no longer need to use a proxy to connect to the Internet, simply set the Proxy type to 'direct' and click the OK button.
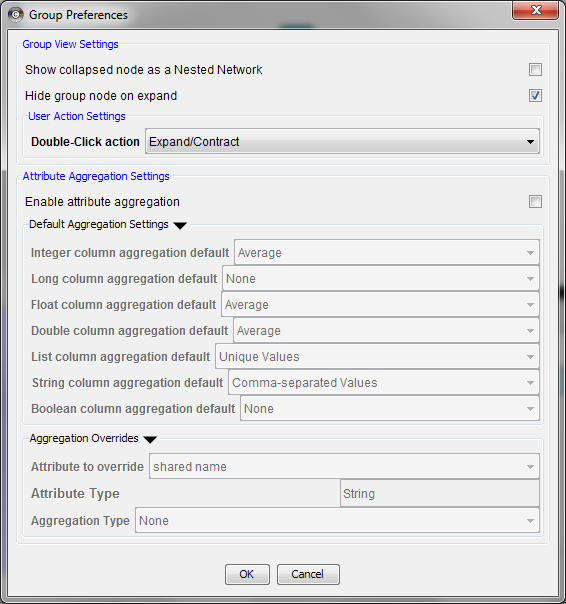
Managing Group view
The configuration of Cytoscape group view may also be edited through Edit → Preferences → Group preferences…
Managing Rendering Engine Settings
Cytoscape's rendering engine uses different thresholds to determine how much detail to use when rendering a graph. When these thresholds are exceeded, the engine stops drawing some features on the graph. The following table summarizes the property names of these thresholds and how they affect rendering:
render.coarseDetailThreshold |
When the number of visible nodes exceeds this value the rendering engine renders nodes and edges with minimal detail. |
render.nodeBorderThreshold |
When the number of visible nodes exceeds this value the rendering engine stops rendering node borders. |
render.nodeLabelThreshold |
When the number of visible nodes exceeds this value the rendering engine stops rendering node labels. |
render.edgeArrowThreshold |
When the number of visible edges exceeds this value the rendering engine stops rendering edge arrows. |
render.edgeLabelThreshold |
When the number of visible edges exceeds this value the rendering engine stops rendering edge labels. |
To force Cytoscape to use full detail always, enable the menu item View → Show Graphics Details.