|
Size: 22066
Comment:
|
Size: 31378
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 1: | Line 1: |
| Interaction networks are useful as stand-alone models. However, they are most powerful for answering scientific questions when integrated with additional information. Cytoscape allows the user to add arbitrary node, edge and network information to Cytoscape as node/edge/network attributes. This could include, for example, annotation data on a gene or confidence values in a protein-protein interaction. These attributes can then be visualized in a user-defined way by setting up a mapping from data attributes to visual attributes (colors, shapes, and so on). The section on visual styles discusses this in greater detail. |
Interaction networks are useful as stand-alone models. However, they are most powerful for answering scientific questions when integrated with additional information. Cytoscape allows the user to add arbitrary node, edge and network data to Cytoscape through the data table. This could include, for example, annotation data on a gene or confidence values in a protein-protein interaction. In the data table information is linked to Nodes, Edges or Networks by mapping them on one of their identifiers. The data can then be visualized in a user-defined way by setting up a mapping from data attributes to visual attributes (colors, shapes, and so on). The section on visual styles discusses this in greater detail. By far the easiest way is by using the txt |
| Line 6: | Line 9: |
|
== Cytoscape Attribute File Format == Node and edge attribute files are simply formatted: a node attribute file begins with the name of the attribute on the first line (note that it cannot contain spaces). Each following line contains the name of the node, followed by an equals sign and the value of that attribute. Numbers and text strings are the most common attribute types. All values for a given attribute must have the same type. For example: {{{ FunctionalCategory YAL001C = metabolism YAR002W = apoptosis YBL007C = ribosome }}} An edge attribute file has much the same structure, except that the name of the edge is the source node name, followed by the interaction type in parentheses, followed by the target node name. Directionality counts, so switching the source and target will refer to a different (or perhaps non-existent) edge. The following is an example edge attributes file: {{{ InteractionStrength YAL001C (pp) YBR043W = 0.82 YMR022W (pd) YDL112C = 0.441 YDL112C (pd) YMR022W = 0.9013 }}} Since Cytoscape treats edge attributes as directional, the second and third edge attribute values refer to two different edges (source and target are reversed, though the nodes involved are the same). Each attribute is stored in a separate file. Node and edge attribute files use the same format. Node and edge attribute file names often use the suffix ".attrs". Node and edge attributes may be loaded at the command line using the –T options or via the File → Import menu. When expression data is loaded using an expression matrix, it is automatically loaded as node attribute data unless explicitly specified otherwise. Node and edge attributes are attached to nodes and edges, and so are independent of networks. Attributes for a given node or edge will be applied to all copies of that node or edge in all loaded network files, regardless of whether the attribute file or network file is imported first. === Detailed file format (Advanced users) === Every attribute file has one header line that gives the name of the attribute, and optionally some additional meta-information about that attribute. The format is as follows: {{{ attributeName (class=JavaClassName) }}} The first field is always the attribute name: it cannot contain spaces. If present, the class field defines the name of the class of the attribute values. For example, java.lang.String or String for Strings, java.lang.Double or Double for floating point values, java.lang.Integer or Integer for integer values, etc. If the value is actually a list of values, the class should be the type of the objects in the list. If no class is specified in the header line, Cytoscape will attempt to guess the type from the first value. If the first value contains numbers in a floating point format, Cytoscape will assume java.lang.Double; if the first value contains only numbers with no decimal point, Cytoscape will assume java.lang.Integer; otherwise Cytoscape will assume java.lang.String. Note that the first value can lead Cytoscape astray: for example, {{{ floatingPointAttribute firstName = 1 secondName = 2.5 }}} In this case, the first value will make Cytoscape think the values should be integers, when in fact they should be floating point numbers. It's safest to explicitly specify the value type to prevent confusion. A better format would be: {{{ floatingPointAttribute (class=Double) firstName = 1 secondName = 2.5 }}} or {{{ floatingPointAttribute firstName = 1.0 secondName = 2.5 }}} Every line past the first line identifies the name of an object (a node in a node attribute file or an edge in a edge attribute file) along with the String representation of the attribute value. The delimiter is always an equals sign; whitespace (spaces and/or tabs) before and after the equals sign is ignored. This means that your names and values can contain whitespace, but object names cannot contain an equals sign and no guarantees are made concerning leading or trailing whitespace. Object names must be the Node ID or Edge ID as seen in the left-most column of the attribute browser if the attribute is to map to anything. These names must be reproduced exactly, including case, or they will not match. Edge names are all of the form: {{{ sourceName (edgeType) targetName }}} Specifically, that is ||sourceName space openParen edgeType closeParen space targetName|| Note that tabs are not allowed in edge names. Tabs can be used to separate the edge name from the "=" delimiter, but not within the edge name itself. Also note that this format is different from the specification of interactions in the SIF file format. To be explicit: a SIF entry for the previous interaction would look like {{{ sourceName edgeType targetName }}} or ||sourceName whiteSpace edgeType whiteSpace targetName|| To specify lists of values, use the following syntax: {{{ listAttributeName (class=java.lang.String) firstObjectName = (firstValue::secondValue::thirdValue) secondObjectName = (onlyOneValue) }}} This example shows an attribute whose value is defined as a list of text strings. The first object has three strings, and thus three elements in its list, while the second object has a list with only one element. In the case of a list every attribute value uses list syntax (i.e. parentheses), and each element is of the same class. Again, the class will be inferred if it is not specified in the header line. Lists are not supported by the visual mapper and so can’t be mapped to visual attributes. === Newline Feature === Sometimes it is desirable to for attributes to include linebreaks, such as node labels that extend over two lines. You can acomplish by inserting {{{\n}}} into the attribute value. For example: {{{ newlineAttr YJL157C = This is a long\nline for a label. }}} == Import Attribute Table Files == In Cytoscape 3.0, importing delimited text and MS Excel attribute data tables is supported. Using this functionality, users can now easily import data that isn't formatted into Cytoscape node or edge attribute file formats (as described above). Also in Cytoscape 3.0, users can select the networks that the imported attributes will be assigned to. In addition to importing attribute files as Node, Edge or Network attributes, it is possible to import unspecified tables which will be shown in the Unassigned Tables tab in table browser. |
== Import Data Table Files == In Cytoscape 3.0, importing delimited text and MS Excel attribute data tables is supported. Using this functionality, users can now easily import data that isn't formatted into Cytoscape node or edge attribute file formats (as described above). Also in Cytoscape 3.0, users can select the networks that the imported attributes will be assigned to. |
| Line 128: | Line 29: |
|
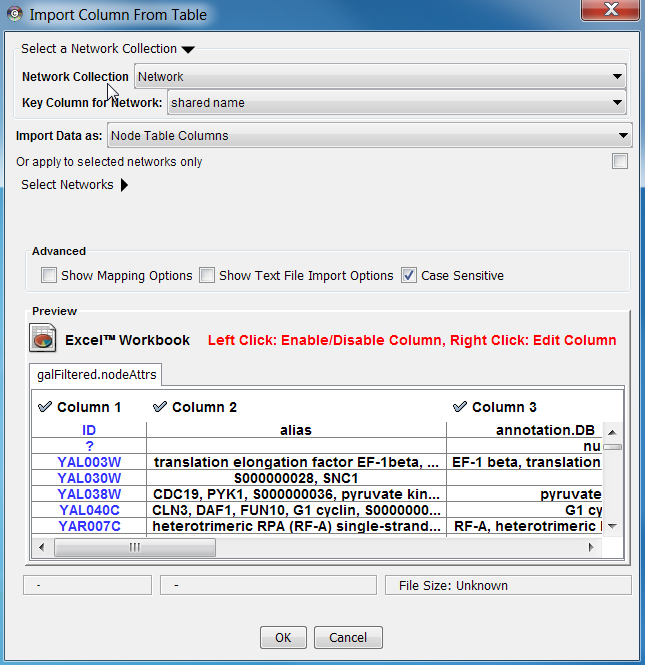
1. In the "Import Attribute from File" panel, select one of the attribute types. Cytoscape can import node, edge, and network attributes as well as unassigned tables.(#1 in figure) 1. (Optional) Choose if you would like to import the file for all of the available networks or only selected networks using the check box in the expandable "Network Options" panel (this panel is collapsed by default). Select networks from the list. (#2 in figure) 1. (Optional) If the table is not properly delimited in the preview panel, change the delimiter in the Text File Import Options panel. The default delimiter is tab. This step is not necessary for Excel Workbooks. (#4 in figure) 1. By default, the first column is designated as the primary key. Change the key column if necessary. (#3 in figure) |
1. In the "Import Attribute from File" panel, select one of the attribute types. Cytoscape can import node, edge, and network attributes. 1. (Optional) Choose if you would like to import the file for all of the available networks or only selected networks using the check box in the expandable "Network Options" panel (this panel is collapsed by default). Select networks from the list. 1. (Optional) If the table is not properly delimited in the preview panel, change the delimiter in the Text File Import Options panel. The default delimiter is tab. This step is not necessary for Excel Workbooks. ) 1. By default, the first column is designated as the primary key. Change the key column if necessary. |
| Line 134: | Line 35: |
|
=== Advanced Options === ==== Mapping Key Attributes to the primary key ==== In Cytoscape 3.0 both IDs and attributes with primitive data types (string, boolean, floating point, and integer) can be selected as the Key Attribute using the dropdown list provided. Complex attributes such as lists are not supported. (#3 in figure) ==== Text File Import Options ==== |
=== Mapping data options === In Cytoscape 3.0 both IDs and attributes with primitive data types (string, boolean, floating point, and integer) can be selected as the Key Attribute using the dropdown list provided. Complex attributes such as lists are not supported. === Data in complete networks === When importing a network from a table other columns than the node ids can be imported as data also. |
| Line 146: | Line 47: |
|
To swap between displaying node, edge, and network attributes use the tabs on the bottom of the panel labeled "Node Attributes", "Edge Attributes", and "Network Attributes". Also, in Cytoscape 3.0 data can be imported which is not assigned to any node, edge or network attributes. This data is displayed in Unassigned Tables tab. In Cytoscape 3.0 there two display modes for table browser: showing only selected nodes/edges attributes or showing all attributes. This configuration can be set using button #1 in the figure. The attribute browser displays attributes belonging to the currently selected network. Using the three buttons under #2 in the figure, it is possible to make some or all columns visible and hide others or all of them. Also, a new column can be created by pressing button #3 or mutable columns can be deleted y button #4. Button #5 is for writing equation which is explained in ????. |
To swap between displaying node, edge, and network attributes use the tabs on the bottom of the panel labeled "Node Attributes", "Edge Attributes", and "Network Attributes". In Cytoscape 3.0 there two display modes for table browser: showing only selected nodes/edges attributes or showing all attributes. This configuration can be set using button (the left most) in the figure. The attribute browser displays attributes belonging to the currently selected network. Using the three buttons (left 2nd to 4th) in the figure, it is possible to make some or all columns visible and hide others or all of them. Also, a new column can be created by pressing button the (left 5th) or mutable columns can be deleted by button (left 6th). Button f(x) is for writing equation which is explained in the section "Supported Functions". |
| Line 154: | Line 55: |
|
Attribute rows in the browser can be sorted alphabetically by specific attribute by clicking on a column heading. A new attribute can be created using the Create New Column button (#3), and must be one of four types – integer, string, real number (floating point), or boolean. Attributes can be deleted using the Delete Attributes button (#4). '''NOTE: Deleting attributes removes them from Cytoscape, not just the attribute browser!''' To remove attributes from the browser without deleting them, simply unselect the attribute using the Select Column button (#2). The right-click menu (#6) on the Table Browser has several functions, such as exporting attribute information to spreadsheet applications. For example, use the right-click menu to Select All and then Copy the data, and then paste it into a spreadsheet application. == Loading Gene Expression (Attribute Matrix) Data == In addition to normal node and edge attribute data, Cytoscape also supports importing gene expression data. Gene expression data are imported using a different file format than normal attributes (File extension is '.pvals'); however, the resulting attributes are not treated differently by Cytoscape. Gene expression data (like attribute data) can be loaded at any time, but are (generally) only relevant once a network has been loaded. === Data File Format === |
Attribute rows in the browser can be sorted alphabetically by specific attribute by clicking on a column heading. A new attribute can be created using the Create New Column button (left 5th), and must be one of four types – integer, string, real number (floating point), or boolean. Attributes can be deleted using the Delete Attributes button (left 6th, trash can icon). '''NOTE: Deleting attributes removes them from Cytoscape, not just the attribute browser!''' To remove attributes from the browser without deleting them, simply unselect the attribute using the Select Column button (left 3rd). The right-click menu on the Table Browser has several functions, such as exporting attribute information to spreadsheet applications. For example, use the right-click menu to Select All and then Copy the data, and then paste it into a spreadsheet application. === Cytoscape Native Data File Formats === Node and edge attribute files are simply formatted: a node attribute file begins with the name of the attribute on the first line (note that it cannot contain spaces). Each following line contains the name of the node, followed by an equals sign and the value of that attribute. Numbers and text strings are the most common attribute types. All values for a given attribute must have the same type. For example: {{{ FunctionalCategory YAL001C = metabolism YAR002W = apoptosis YBL007C = ribosome }}} An edge attribute file has much the same structure, except that the name of the edge is the source node name, followed by the interaction type in parentheses, followed by the target node name. Directionality counts, so switching the source and target will refer to a different (or perhaps non-existent) edge. The following is an example edge attributes file: {{{ InteractionStrength YAL001C (pp) YBR043W = 0.82 YMR022W (pd) YDL112C = 0.441 YDL112C (pd) YMR022W = 0.9013 }}} Since Cytoscape treats edge attributes as directional, the second and third edge attribute values refer to two different edges (source and target are reversed, though the nodes involved are the same). Each attribute is stored in a separate file. Node and edge attribute files use the same format. Node and edge attribute file names often use the suffix ".attrs". Node and edge attributes may be loaded at the command line using the –T options or via the File → Import menu. When expression data is loaded using an expression matrix, it is automatically loaded as node attribute data unless explicitly specified otherwise. Node and edge attributes are attached to nodes and edges, and so are independent of networks. Attributes for a given node or edge will be applied to all copies of that node or edge in all loaded network files, regardless of whether the attribute file or network file is imported first. === Detailed file format (Advanced users) === Every attribute file has one header line that gives the name of the attribute, and optionally some additional meta-information about that attribute. The format is as follows: {{{ attributeName (class=JavaClassName) }}} The first field is always the attribute name: it cannot contain spaces. If present, the class field defines the name of the class of the attribute values. For example, java.lang.String or String for Strings, java.lang.Double or Double for floating point values, java.lang.Integer or Integer for integer values, etc. If the value is actually a list of values, the class should be the type of the objects in the list. If no class is specified in the header line, Cytoscape will attempt to guess the type from the first value. If the first value contains numbers in a floating point format, Cytoscape will assume java.lang.Double; if the first value contains only numbers with no decimal point, Cytoscape will assume java.lang.Integer; otherwise Cytoscape will assume java.lang.String. Note that the first value can lead Cytoscape astray: for example, {{{ floatingPointAttribute firstName = 1 secondName = 2.5 }}} In this case, the first value will make Cytoscape think the values should be integers, when in fact they should be floating point numbers. It's safest to explicitly specify the value type to prevent confusion. A better format would be: {{{ floatingPointAttribute (class=Double) firstName = 1 secondName = 2.5 }}} or {{{ floatingPointAttribute firstName = 1.0 secondName = 2.5 }}} Every line past the first line identifies the name of an object (a node in a node attribute file or an edge in a edge attribute file) along with the String representation of the attribute value. The delimiter is always an equals sign; whitespace (spaces and/or tabs) before and after the equals sign is ignored. This means that your names and values can contain whitespace, but object names cannot contain an equals sign and no guarantees are made concerning leading or trailing whitespace. Object names must be the Node ID or Edge ID as seen in the left-most column of the attribute browser if the attribute is to map to anything. These names must be reproduced exactly, including case, or they will not match. Edge names are all of the form: {{{ sourceName (edgeType) targetName }}} Specifically, that is ||sourceName space openParen edgeType closeParen space targetName|| Note that tabs are not allowed in edge names. Tabs can be used to separate the edge name from the "=" delimiter, but not within the edge name itself. Also note that this format is different from the specification of interactions in the SIF file format. To be explicit: a SIF entry for the previous interaction would look like {{{ sourceName edgeType targetName }}} or ||sourceName whiteSpace edgeType whiteSpace targetName|| To specify lists of values, use the following syntax: {{{ listAttributeName (class=java.lang.String) firstObjectName = (firstValue::secondValue::thirdValue) secondObjectName = (onlyOneValue) }}} This example shows an attribute whose value is defined as a list of text strings. The first object has three strings, and thus three elements in its list, while the second object has a list with only one element. In the case of a list every attribute value uses list syntax (i.e. parentheses), and each element is of the same class. Again, the class will be inferred if it is not specified in the header line. Lists are not supported by the visual mapper and so can’t be mapped to visual attributes. ==== Newline Feature ==== Sometimes it is desirable to for attributes to include linebreaks, such as node labels that extend over two lines. You can acomplish by inserting {{{\n}}} into the attribute value. For example: {{{ newlineAttr YJL157C = This is a long\nline for a label. }}} === Gene Expression (Attribute Matrix) Data format === In addition to normal node and edge attribute data, Cytoscape also supports importing gene expression data. Gene expression data are imported using a different file format than normal attributes (File extension is '.pvals'); however, the resulting attributes are not treated differently by Cytoscape. ==== Data File Format ==== |
| Line 193: | Line 195: |
|
Some variations on this basic format are recognized; see the formal file format specification below for more information. Expression data files commonly have the file extension ".pvals", and this file extension is recognized by Cytoscape when browsing for data files (File->Import->Table->File...). === General Procedure === Load an expression attribute matrix file using File → Import → Attribute/Expression Matrix... to bring up the import window, or by specifying the filename using the -m option at the command line. If you use the command line input, you must enter your expression data by node ID. If you use the dialog box, then you can either load expression data by node ID (the default option), or you can select a node attribute to use in assigning your expression data to your Cytoscape nodes. If you do use a node attribute, then (1) the attribute should already be loaded, and (2) the node attribute value must match the first column in your matrix file. ==== Worked Example ==== |
Some variations on this basic format are recognized; see the formal file format specification below for more information. Expression data files commonly have the file extension ".pvals", and this file extension is recognized by Cytoscape when browsing for data files (File → Import → Table → File...). ==== General Procedure ==== Load an expression attribute matrix file using File → Import → Table → File... to bring up the import window, or by specifying the filename using the -m option at the command line. ===== Worked Example ===== |
| Line 202: | Line 203: |
|
'''Option A.''' Load a sample gene expression data set by going to File → Import → Attribute/Expression Matrix... . In the resulting window, in the field labeled "Please select an attribute or expression matrix file...", use the Select button to enter {{{sampleData/galExpData.pvals}}}. The identifiers used in this file are the same ones used in the network file {{{sampleData/galFiltered.sif}}}, so you do not need to touch the field labeled "Assign values to nodes using...". A few lines of this file are shown below: |
Load a sample gene expression data set by going to File → Import → Table → File.... In the resulting window, select the file {{{sampleData/galExpData.pvals}}}. In the combobox 'Import Data to', make sure the selected item is 'Node Attributes'. The identifiers used in this file are the same ones used in the network file {{{sampleData/galFiltered.sif}}}, so the attributes will be mapped to the nodes automatically. A few lines of this file are shown below: |
| Line 213: | Line 212: |
|
'''Option B.''' Step 1. After loading the network, load the node attribute file {{{sampleData/gal.probeset.na}}}, using File → Import → Node attributes... . This file is shown in part below: {{{ Probeset YHR051W = probeset2 YHR124W = probeset3 YKL181W = probeset4 }}} Step 2. After loading the node attribute file, select the expression data file {{{sampleData.galExpPvals.probeset.pvals}}}, shown in part below: {{{ GENE COMMON gal1RG gal4RG gal80R gal1RG gal4RG gal80R probeset2 COX6 -0.034 0.111 -0.304 3.75720e-01 1.56240e-02 7.91340e-06 probeset3 NDT80 -0.090 0.007 -0.348 2.71460e-01 9.64330e-01 3.44760e-01 probeset4 PRS1 -0.167 -0.233 0.112 6.27120e-03 7.89400e-04 1.44060e-01 }}} After selecting this file, in the field labeled "Assign values to nodes using...", select Probeset. You will see that this loads exactly the same expression data as in Case 1, but provides extra flexibility in case the node name cannot be used as an identifier. ==== Detailed file format (Advanced users) ==== In all expression data files, any whitespace (spaces and/or tabs) is considered a delimiter between adjacent fields. Every line of text is either the header line or contains all the measurements for a particular gene. No name conversion is applied to expression data files. |
===== Detailed file format (Advanced users) ===== Expression data files must have extension '.pvals'. In all expression data files, any whitespace (spaces and/or tabs) is considered a delimiter between adjacent fields. Every line of text is either the header line or contains all the measurements for a particular gene. No name conversion is applied to expression data files. |
| Line 267: | Line 245: |
|
== Annotations == Annotations in Cytoscape are stored as a set of ontologies (e.g. the Gene Ontology, or GO). An ontology consists of a set of controlled vocabulary terms that annotate the objects. For example, using the Gene Ontology, the Saccharomyces Cerevisiae '''CDC55''' gene has a biological process described as “protein biosynthesis”, to which GO has assigned the number 6412 (a GO ID). {{{ GO 8150 biological_process GO 7582 physiological processes GO 8152 metabolism GO 44238 primary metabolism GO 19538 protein metabolism GO 6412 protein biosynthesis }}} '''Graphical View of GO Term 6412: protein biosynthesis''' {{attachment:ontology_dag1.png}} Cytoscape can use this ontology DAG (Directed Acyclic Graph) to annotate objects in networks. The Ontology Server is a Cytoscape feature which allows you to load, navigate, and assign annotation terms to nodes and edges in a network. Cytoscape has an GUI for loading ontology and associated annotation, enabling you to load both local and remote files. === Ontology and Annotation File Format === The standard file formats used in Cytoscape Ontology Server are OBO and Gene Association. The GO website details these file formats: * Ontologies and Definitions: http://www.geneontology.org/GO.downloads.shtml#ont * Current Annotations: http://www.geneontology.org/GO.current.annotations.shtml ==== OBO File ==== An OBO file is the ontology DAG itself. This file defines the relationships between ontology terms. Cytoscape can load all ontology files written in OBO format. The full listing of ontology files are available from the Open Biomedical Ontologies (OBO) website: * OBO Ontology Browser: http://obo.sourceforge.net/browse.html Sample OBO File - gene_ontology.obo: http://www.geneontology.org/ontology/gene_ontology_edit.obo {{{ format-version: 1.2 date: 27:11:2006 17:12 saved-by: midori auto-generated-by: OBO-Edit 1.002 subsetdef: goslim_generic "Generic GO slim" subsetdef: goslim_goa "GOA and proteome slim" subsetdef: goslim_plant "Plant GO slim" subsetdef: goslim_yeast "Yeast GO slim" subsetdef: gosubset_prok "Prokaryotic GO subset" default-namespace: gene_ontology remark: cvs version: $Revision: 5.49 $ [Term] id: GO:0000001 name: mitochondrion inheritance namespace: biological_process def: "The distribution of mitochondria, including the mitochondrial genome, into daughter cells after mitosis or meiosis, mediated by interactions between mitochondria and the cytoskeleton." [GOC:mcc, PMID:10873824, PMID:11389764] synonym: "mitochondrial inheritance" EXACT [] is_a: GO:0048308 ! organelle inheritance is_a: GO:0048311 ! mitochondrion distribution [Term] id: GO:0000002 name: mitochondrial genome maintenance namespace: biological_process def: "The maintenance of the structure and integrity of the mitochondrial genome." [GOC:ai] is_a: GO:0007005 ! mitochondrion organization and biogenesis }}} ==== Default List of Ontologies ==== Cytoscape provides a list of ontologies available in OBO format. If an Internet connection is available, Cytoscape will import ontology and annotation files directly from the remote source. The table below lists the included ontologies. || '''Ontology Name''' || '''Description''' || ||Gene Ontology Full||This data source contains a full-size GO DAG, which contains all GO terms. This OBO file is written in version 1.2 format.|| ||Generic GO slim|| A subset of general GO Terms, including higer-level terms only.|| ||Yeast GO slim||A subset of GO Terms for annotating Yeast data sets maintained by SGD.|| ||Molecule role (INOH Protein name/family name ontology)||A structured controlled vocabulary of concrete and abstract (generic) protein names. This ontology is a INOH pathway annotation ontology, one of a set of ontologies intended to be used in pathway data annotation to ease data integration. This ontology is used to annotate protein names, protein family names, and generic/concrete protein names in the INOH pathway data. INOH is part of the BioPAX working group.|| ||Event (INOH pathway ontology)||A structured controlled vocabulary of pathway-centric biological processes. This ontology is a INOH pathway annotation ontology, one of a set of ontologies intended to be used in pathway data annotation to ease data integration. This ontology is used to annotate biological processes, pathways, and sub-pathways in the INOH pathway data. INOH is part of the BioPAX working group.|| ||Protein-protein interaction||A structured controlled vocabulary for the annotation of experiments concerned with protein-protein interactions.|| ||Pathway Ontology||The Pathway Ontology is a controlled vocabulary for pathways that provides standard terms for the annotation of gene products.|| ||PATO||PATO is an ontology of phenotypic qualities, intended for use in a number of applications, primarily phenotype annotation. For more information, please visit the PATO wiki (http://www.bioontology.org/wiki/index.php/PATO:Main_Page). || ||Mouse pathology||The Mouse Pathology Ontology (MPATH) is an ontology for mutant mouse pathology. This is Version 1.|| ||Human disease||This ontology is a comprehensive hierarchical controlled vocabulary for human disease representation. For more information, please visit the Disease Ontology website (http://diseaseontology.sourceforge.net/).|| Although Cytoscape can import all kinds of ontologies in OBO format, annotation files are associated with specific ontologies. Therefore, you need to provide the correct ontology-specific annotation file to annotate nodes/edges/networks in Cytoscape. For example, while you can annotate human network data using the GO Full ontology with human Gene Association files, you cannot use a combination of the human Disease Ontology file and human Gene Association files, because the Gene Association file is only compatible with GO. ==== Visualize and Browse Ontology DAG (for Advanced Users) ==== Relationships between ontology terms are usually represented as Directed Acyclic Graphs (DAGs). This is a special case of a network (or graph), where nodes are ontology terms and edges are relationships between terms. Ontology data is stored in the same data structure as normal networks. This enables users and App writers to visualize, browse and manipulate ontology DAGs just like other networks. The following is an example of visualization of an ontology DAG (Generic GO Slim): {{attachment:ontology_dag2.png}} Every ontology term and relationship can have attributes. In the OBO files, ontology terms have optional fields such as definition, synonyms, comments, or cross-references. These fields will be imported as node attributes. To browse those attributes, please use the attribute browser (see the example below): {{attachment:ontology_attrs.png}} * Note 1: Some ontologies have a lot of terms. For example, the full Gene Ontology contains more than 20,000 terms. If you need to save memory, you can remove this ontology DAG from Network Panel (right-click on the ontology name at the left-hand side of the screen and select Destroy Network). * Note 2: All ontology DAGs will be saved in the session file. To minimize the session file size, you can delete the Ontology DAG before saving session. ==== Gene Association File ==== The Gene Association (GA) file provides annotation only for the Gene Ontology. It is a species-specific annotation file for GO terms. '''Gene Association files will only work with Gene Ontologies and NOT others!''' Sample Gene Association File ([[http://viewvc.geneontology.org/viewvc/GO-SVN/trunk/gene-associations/gene_association.sgd.gz?rev=HEAD|gene_association.sgd]] - annotation file for yeast): {{{ SGD S000003916 AAD10 GO:0006081 SGD_REF:S000042151|PMID:10572264 ISS P aryl-alcohol dehydrogenase (putative) YJR155W gene taxon:4932 20020902 SGD SGD S000005275 AAD14 GO:0008372 SGD_REF:S000069584 ND C aryl-alcohol dehydrogenase (putative) YNL331C gene taxon:4932 20010119 SGD }}} === Node Name Mapping === If you have a network file and an attribute file, they should have a common key to map attributes onto network data. If those two do not have a common key, you need to do an extra step to add a shared key. The following is a quick tutorial to learn how to use Gene Name Mapping files. ==== Import New ID Sets from BioMart ==== You can import various kinds of ID sets from !BioMart (http://www.biomart.org/index.html). !BioMart web service client is integrated into Cytoscape. {{attachment:id_mapping1.png}} 1. Select: '''File → Import → Table → Public Databases...''' 1. Select a data source. For ID mapping, select one of the ''Ensemble Genes'' data set. You need to choose correct species for your network. 1. Select '''Attribute'''. If you want to import new ID sets matching current node IDs, select ''name''. 1. Select '''Data Type'''. This should be the type of ID set selected in '''Attribute''' list. For example, if you select ''name'' for '''Attribute''' and your network uses ''Entrez Gene ID'' for its node ID, you need to select ''!EntrezGene ID(s)'' for '''Data Type'''. 1. Select new ID sets from the list. Because !BioMart server does not accept query to import lots of annotations at once, you can select only 3-5 attributes for each import. 1. Press '''Import'''. ==== Import Gene Ontology and Gene Association Files ==== For convenience, Cytoscape has a list of URLs for commonly used ontology data and a complete set of Gene Association files. To import Gene Ontology and Gene Association files for the loaded networks, please follow these steps: '''''Important: All data sources in the preset list are remote URLs, meaning a network connection is required!''''' ===== Step 1. Select an Annotation File ===== {{attachment:ontology_import_annotation.png}} Select File → Import → Ontology and Annotation... to open the "Import Ontology and Annotation" window. From the Annotation dropdown list, select a gene association file for your network. For example, if you want to annotate the yeast network, select "Gene Association file for Saccharomyces cerevisiae". ===== Step 2. Select an Ontology File ===== {{attachment:ontology_import_obo.png}} Select an Ontology data (OBO file) from the Ontology dropdown list. If the file is not loaded yet, it will be shown in red. The first three files are Gene Ontology files. You can load other ontologies, but you need your own annotation file to annotate networks. ===== Step 3. Import the files ===== Once you click the Import button, Cytoscape will start loading OBO and Gene Association files from the remote sources. If you choose GO Full it may take a while since it is a large data file. |
Interaction networks are useful as stand-alone models. However, they are most powerful for answering scientific questions when integrated with additional information. Cytoscape allows the user to add arbitrary node, edge and network data to Cytoscape through the data table. This could include, for example, annotation data on a gene or confidence values in a protein-protein interaction. In the data table information is linked to Nodes, Edges or Networks by mapping them on one of their identifiers. The data can then be visualized in a user-defined way by setting up a mapping from data attributes to visual attributes (colors, shapes, and so on). The section on visual styles discusses this in greater detail.
By far the easiest way is by using the txt
Cytoscape supports the importing of attribute in some predefined format. Cytoscape can find the right reader based on the file extension of attribute file. The supported file extensions are "attrs" (text file in the format described below), "tsv, tab, csv, net or txt" (comma or tab separated values file), "pvals" (Cytoscape expression matrix) and "xls, xlsx" (Microsost Excel file).
Import Data Table Files
In Cytoscape 3.0, importing delimited text and MS Excel attribute data tables is supported. Using this functionality, users can now easily import data that isn't formatted into Cytoscape node or edge attribute file formats (as described above). Also in Cytoscape 3.0, users can select the networks that the imported attributes will be assigned to.
Sample Attribute Table 1
Object Key |
SGD ID |
AAC3 |
S000000289 |
AAT2 |
S000004017 |
BIK1 |
S000000534 |
The attribute table file should contain a primary key column and at least one attribute column. The maximum number of attribute columns is unlimited. Alternatively, you can specify each attribute name from the File → Import → Attribute from Table (text/MS Excel)... user interface.
Basic Operation
The user interface of the "Import Attributes from Table" window is similar to that of the "Import Network from Table" window.
Select File → Import → Table → File... ( or URL... if your source data file is accessible through web)
- Select a data file in the file chooser panel (or enter the URL in the displayed box). This file can be either a text or Excel (.xls) formats.
- In the "Import Attribute from File" panel, select one of the attribute types. Cytoscape can import node, edge, and network attributes.
- (Optional) Choose if you would like to import the file for all of the available networks or only selected networks using the check box in the expandable "Network Options" panel (this panel is collapsed by default). Select networks from the list.
- (Optional) If the table is not properly delimited in the preview panel, change the delimiter in the Text File Import Options panel. The default delimiter is tab. This step is not necessary for Excel Workbooks. )
- By default, the first column is designated as the primary key. Change the key column if necessary.
- Click the Import button.
Mapping data options
In Cytoscape 3.0 both IDs and attributes with primitive data types (string, boolean, floating point, and integer) can be selected as the Key Attribute using the dropdown list provided. Complex attributes such as lists are not supported.
Data in complete networks
When importing a network from a table other columns than the node ids can be imported as data also. For more detail on these options, please see the "Import Free-Format Table Files" section of the user manual in the Creating Networks chapter.
Table Browser
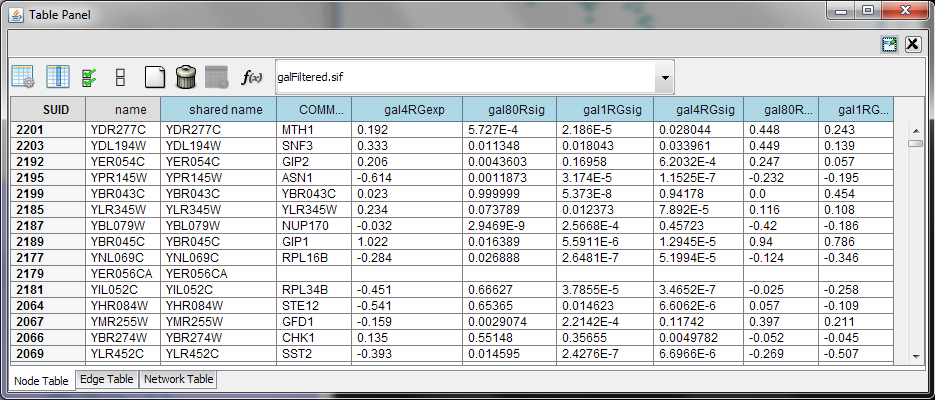
When Cytoscape is started, the Table Browser appears in the bottom CytoPanel. This browser can be hidden and restored using the View → Show/Hide Table Browser menu option. Like other CytoPanels, the browser can be undocked by pressing the little icon in the browser’s top right corner.
To swap between displaying node, edge, and network attributes use the tabs on the bottom of the panel labeled "Node Attributes", "Edge Attributes", and "Network Attributes".
In Cytoscape 3.0 there two display modes for table browser: showing only selected nodes/edges attributes or showing all attributes. This configuration can be set using button (the left most) in the figure. The attribute browser displays attributes belonging to the currently selected network.
Using the three buttons (left 2nd to 4th) in the figure, it is possible to make some or all columns visible and hide others or all of them. Also, a new column can be created by pressing button the (left 5th) or mutable columns can be deleted by button (left 6th). Button f(x) is for writing equation which is explained in the section "Supported Functions".
Most attribute values can be edited by double-clicking an attribute cell (only the SUID cannot be edited). Newline characters can be inserted into String attributes either by pressing Enter or by typing "\n". Once finished editing, click outside of the editing cell in the Attribute Browser or press Shift-Enter to save your edits. Pressing Esc while editing will undo any changes.
Attribute rows in the browser can be sorted alphabetically by specific attribute by clicking on a column heading. A new attribute can be created using the Create New Column button (left 5th), and must be one of four types – integer, string, real number (floating point), or boolean. Attributes can be deleted using the Delete Attributes button (left 6th, trash can icon). NOTE: Deleting attributes removes them from Cytoscape, not just the attribute browser! To remove attributes from the browser without deleting them, simply unselect the attribute using the Select Column button (left 3rd).
The right-click menu on the Table Browser has several functions, such as exporting attribute information to spreadsheet applications. For example, use the right-click menu to Select All and then Copy the data, and then paste it into a spreadsheet application.
Cytoscape Native Data File Formats
Node and edge attribute files are simply formatted: a node attribute file begins with the name of the attribute on the first line (note that it cannot contain spaces). Each following line contains the name of the node, followed by an equals sign and the value of that attribute. Numbers and text strings are the most common attribute types. All values for a given attribute must have the same type. For example:
FunctionalCategory YAL001C = metabolism YAR002W = apoptosis YBL007C = ribosome
An edge attribute file has much the same structure, except that the name of the edge is the source node name, followed by the interaction type in parentheses, followed by the target node name. Directionality counts, so switching the source and target will refer to a different (or perhaps non-existent) edge. The following is an example edge attributes file:
InteractionStrength YAL001C (pp) YBR043W = 0.82 YMR022W (pd) YDL112C = 0.441 YDL112C (pd) YMR022W = 0.9013
Since Cytoscape treats edge attributes as directional, the second and third edge attribute values refer to two different edges (source and target are reversed, though the nodes involved are the same).
Each attribute is stored in a separate file. Node and edge attribute files use the same format. Node and edge attribute file names often use the suffix ".attrs".
Node and edge attributes may be loaded at the command line using the –T options or via the File → Import menu.
When expression data is loaded using an expression matrix, it is automatically loaded as node attribute data unless explicitly specified otherwise.
Node and edge attributes are attached to nodes and edges, and so are independent of networks. Attributes for a given node or edge will be applied to all copies of that node or edge in all loaded network files, regardless of whether the attribute file or network file is imported first.
Detailed file format (Advanced users)
Every attribute file has one header line that gives the name of the attribute, and optionally some additional meta-information about that attribute. The format is as follows:
attributeName (class=JavaClassName)
The first field is always the attribute name: it cannot contain spaces. If present, the class field defines the name of the class of the attribute values. For example, java.lang.String or String for Strings, java.lang.Double or Double for floating point values, java.lang.Integer or Integer for integer values, etc. If the value is actually a list of values, the class should be the type of the objects in the list. If no class is specified in the header line, Cytoscape will attempt to guess the type from the first value. If the first value contains numbers in a floating point format, Cytoscape will assume java.lang.Double; if the first value contains only numbers with no decimal point, Cytoscape will assume java.lang.Integer; otherwise Cytoscape will assume java.lang.String. Note that the first value can lead Cytoscape astray: for example,
floatingPointAttribute firstName = 1 secondName = 2.5
In this case, the first value will make Cytoscape think the values should be integers, when in fact they should be floating point numbers. It's safest to explicitly specify the value type to prevent confusion. A better format would be:
floatingPointAttribute (class=Double) firstName = 1 secondName = 2.5
or
floatingPointAttribute firstName = 1.0 secondName = 2.5
Every line past the first line identifies the name of an object (a node in a node attribute file or an edge in a edge attribute file) along with the String representation of the attribute value. The delimiter is always an equals sign; whitespace (spaces and/or tabs) before and after the equals sign is ignored. This means that your names and values can contain whitespace, but object names cannot contain an equals sign and no guarantees are made concerning leading or trailing whitespace. Object names must be the Node ID or Edge ID as seen in the left-most column of the attribute browser if the attribute is to map to anything. These names must be reproduced exactly, including case, or they will not match.
Edge names are all of the form:
sourceName (edgeType) targetName
Specifically, that is
sourceName space openParen edgeType closeParen space targetName |
Note that tabs are not allowed in edge names. Tabs can be used to separate the edge name from the "=" delimiter, but not within the edge name itself. Also note that this format is different from the specification of interactions in the SIF file format. To be explicit: a SIF entry for the previous interaction would look like
sourceName edgeType targetName
or
sourceName whiteSpace edgeType whiteSpace targetName |
To specify lists of values, use the following syntax:
listAttributeName (class=java.lang.String) firstObjectName = (firstValue::secondValue::thirdValue) secondObjectName = (onlyOneValue)
This example shows an attribute whose value is defined as a list of text strings. The first object has three strings, and thus three elements in its list, while the second object has a list with only one element. In the case of a list every attribute value uses list syntax (i.e. parentheses), and each element is of the same class. Again, the class will be inferred if it is not specified in the header line. Lists are not supported by the visual mapper and so can’t be mapped to visual attributes.
Newline Feature
Sometimes it is desirable to for attributes to include linebreaks, such as node labels that extend over two lines. You can acomplish by inserting \n into the attribute value. For example:
newlineAttr YJL157C = This is a long\nline for a label.
Gene Expression (Attribute Matrix) Data format
In addition to normal node and edge attribute data, Cytoscape also supports importing gene expression data. Gene expression data are imported using a different file format than normal attributes (File extension is '.pvals'); however, the resulting attributes are not treated differently by Cytoscape.
Data File Format
Gene expression ratios or values are specified over one or more experiments using a text file. Ratios result from a comparison of two expression measurements (experiment vs. control). Some expression platforms, such as Affymetrix, directly measure expression values, without a comparison. The file consists of a header and a number of space- or tab-delimited fields, one line per gene, with the following format:
Identifier [CommonName] value1 value2 ... valueN [pval1 pval2 ... pvalN]
Brackets [ ] indicate fields that are optional.
The first field identifies which Cytoscape node the data refers to. In the simplest case, this is the gene name - exactly as it appears on the network generated by Cytoscape (case sensitive!). Alternatively, this can be some node attribute that identifies the node uniquely, such as a probeset identifier for commercial microarrays.
The next field is an optional common name. It is not used by Cytoscape, and is provided strictly for the user's convenience. With this common name field, the input format is the same as for commonly-used expression data anaysis packages such as SAM (http://www-stat.stanford.edu/~tibs/SAM/).
The next set of columns represent expression values, one per experiment. These can be either absolute expression values or fold change ratios. Each experiment is identified by its experiment name, given in the first line.
Optionally, significance measures such as P values may be provided. These values, generated by many microarray data analysis packages, indicate where the level of gene expression or the fold change appears to be greater than random chance. If you are using significance measures, then your expression file should contain them in a second set of columns after the expression values. The column names for the expression significance measures need to match those of the expression values exactly.
For example, here is an excerpt from the file galExpData.pvals in the Cytoscape sampleData directory:
GENE COMMON gal1RG gal4RG gal80R gal1RG gal4RG gal80R YHR051W COX6 -0.034 0.111 -0.304 3.75720e-01 1.56240e-02 7.91340e-06 YHR124W NDT80 -0.090 0.007 -0.348 2.71460e-01 9.64330e-01 3.44760e-01 YKL181W PRS1 -0.167 -0.233 0.112 6.27120e-03 7.89400e-04 1.44060e-01 YGR072W UPF3 0.245 -0.471 0.787 4.10450e-04 7.51780e-04 1.37130e-05
This indicates that there is data for three experiments: gal1RG, gal4RG, and gal80R. These names appear two times in the header line: the first time gives the expression values, and the second gives the significance measures. For instance, the second line tells us that in Experiment gal1RG, the gene YHR051W has an expression value of -0.034 with significance measure 3.75720e-01.
Some variations on this basic format are recognized; see the formal file format specification below for more information. Expression data files commonly have the file extension ".pvals", and this file extension is recognized by Cytoscape when browsing for data files (File → Import → Table → File...).
General Procedure
Load an expression attribute matrix file using File → Import → Table → File... to bring up the import window, or by specifying the filename using the -m option at the command line.
Worked Example
For the sample network file sampleData/galFiltered.sif:
Load a sample gene expression data set by going to File → Import → Table → File.... In the resulting window, select the file sampleData/galExpData.pvals. In the combobox 'Import Data to', make sure the selected item is 'Node Attributes'. The identifiers used in this file are the same ones used in the network file sampleData/galFiltered.sif, so the attributes will be mapped to the nodes automatically. A few lines of this file are shown below:
GENE COMMON gal1RG gal4RG gal80R gal1RG gal4RG gal80R YHR051W COX6 -0.034 0.111 -0.304 3.75720e-01 1.56240e-02 7.91340e-06 YHR124W NDT80 -0.090 0.007 -0.348 2.71460e-01 9.64330e-01 3.44760e-01 YKL181W PRS1 -0.167 -0.233 0.112 6.27120e-03 7.89400e-04 1.44060e-01
Detailed file format (Advanced users)
Expression data files must have extension '.pvals'. In all expression data files, any whitespace (spaces and/or tabs) is considered a delimiter between adjacent fields. Every line of text is either the header line or contains all the measurements for a particular gene. No name conversion is applied to expression data files.
The names given in the first column of the expression data file should match exactly the names used elsewhere (i.e. in SIF or GML files).
The first line is a header line with one of the following three header formats:
<text> <text> cond1 cond2 ... cond1 cond2 ... [NumSigConds] <text> <text> cond1 cond2 ... <tab><tab>RATIOS<tab><tab>...LAMBDAS
The first format specifies that both expression ratios and significance values are included in the file. The first two text tokens (in angled brackets) contain names for each gene, such as the formal and common gene names. The condX token set specifies the names of the experimental conditions; these columns will contain ratio values. This list of condition names must then be duplicated exactly, each spelled the same way and in the same order. Optionally, a final column with the title NumSigConds may be present. If present, this column will contain integer values indicating the number of conditions in which each gene had a statistically significant change according to some threshold.
The second format is similar to the first except that the duplicate column names are omitted, and there is no NumSigConds field. This format specifies data with ratios but no significance values.
The third format specifies an MTX header, which is a commonly used format. Two tab characters precede the RATIOS token. This token is followed by a number of tabs equal to the number of conditions, followed by the LAMBDAS token. This format specifies both ratios and significance values.
Each line after the first is a data line with the following format:
FormalGeneName CommonGeneName ratio1 ratio2 ... [lambda1 lambda2 ...] [numSigConds]
The first two tokens are gene names. The names in the first column are the keys used for node name lookup; these names should be the same as the names used elsewhere in Cytoscape (i.e. in the SIF, GML, or XGMML files). Traditionally in the gene expression microarray community, who defined these file formats, the first token is expected to be the formal name of the gene (in systems where there is a formal naming scheme for genes), while the second is expected to be a synonym for the gene commonly used by biologists, although Cytoscape does not make use of the common name column. The next columns contain floating point values for the ratios, followed by columns with the significance values if specified by the header line. The final column, if specified by the header line, should contain an integer giving the number of significant conditions for that gene. Missing values are not allowed and will confuse the parser. For example, using two consecutive tabs to indicate a missing value will not work; the parser will regard both tabs as a single delimiter and be unable to parse the line correctly.
Optionally, the last line of the file may be a special footer line with the following format:
NumSigGenes int1 int2 ...
This line specified the number of genes that were significantly differentially expressed in each condition. The first text token must be spelled exactly as shown; the rest of the line should contain one integer value for each experimental condition.
Annotations
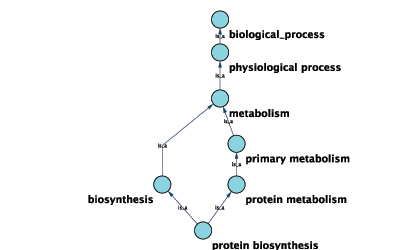
Annotations in Cytoscape are stored as a set of ontologies (e.g. the Gene Ontology, or GO). An ontology consists of a set of controlled vocabulary terms that annotate the objects. For example, using the Gene Ontology, the Saccharomyces Cerevisiae CDC55 gene has a biological process described as “protein biosynthesis”, to which GO has assigned the number 6412 (a GO ID).
GO 8150 biological_process
GO 7582 physiological processes
GO 8152 metabolism
GO 44238 primary metabolism
GO 19538 protein metabolism
GO 6412 protein biosynthesisGraphical View of GO Term 6412: protein biosynthesis
Cytoscape can use this ontology DAG (Directed Acyclic Graph) to annotate objects in networks. The Ontology Server is a Cytoscape feature which allows you to load, navigate, and assign annotation terms to nodes and edges in a network. Cytoscape has an GUI for loading ontology and associated annotation, enabling you to load both local and remote files.
Ontology and Annotation File Format
The standard file formats used in Cytoscape Ontology Server are OBO and Gene Association. The GO website details these file formats:
Ontologies and Definitions: http://www.geneontology.org/GO.downloads.shtml#ont
Current Annotations: http://www.geneontology.org/GO.current.annotations.shtml
OBO File
An OBO file is the ontology DAG itself. This file defines the relationships between ontology terms. Cytoscape can load all ontology files written in OBO format. The full listing of ontology files are available from the Open Biomedical Ontologies (OBO) website:
OBO Ontology Browser: http://obo.sourceforge.net/browse.html
Sample OBO File - gene_ontology.obo: http://www.geneontology.org/ontology/gene_ontology_edit.obo
format-version: 1.2 date: 27:11:2006 17:12 saved-by: midori auto-generated-by: OBO-Edit 1.002 subsetdef: goslim_generic "Generic GO slim" subsetdef: goslim_goa "GOA and proteome slim" subsetdef: goslim_plant "Plant GO slim" subsetdef: goslim_yeast "Yeast GO slim" subsetdef: gosubset_prok "Prokaryotic GO subset" default-namespace: gene_ontology remark: cvs version: $Revision: 5.49 $ [Term] id: GO:0000001 name: mitochondrion inheritance namespace: biological_process def: "The distribution of mitochondria, including the mitochondrial genome, into daughter cells after mitosis or meiosis, mediated by interactions between mitochondria and the cytoskeleton." [GOC:mcc, PMID:10873824, PMID:11389764] synonym: "mitochondrial inheritance" EXACT [] is_a: GO:0048308 ! organelle inheritance is_a: GO:0048311 ! mitochondrion distribution [Term] id: GO:0000002 name: mitochondrial genome maintenance namespace: biological_process def: "The maintenance of the structure and integrity of the mitochondrial genome." [GOC:ai] is_a: GO:0007005 ! mitochondrion organization and biogenesis
Default List of Ontologies
Cytoscape provides a list of ontologies available in OBO format. If an Internet connection is available, Cytoscape will import ontology and annotation files directly from the remote source. The table below lists the included ontologies.
Ontology Name |
Description |
Gene Ontology Full |
This data source contains a full-size GO DAG, which contains all GO terms. This OBO file is written in version 1.2 format. |
Generic GO slim |
A subset of general GO Terms, including higer-level terms only. |
Yeast GO slim |
A subset of GO Terms for annotating Yeast data sets maintained by SGD. |
Molecule role (INOH Protein name/family name ontology) |
A structured controlled vocabulary of concrete and abstract (generic) protein names. This ontology is a INOH pathway annotation ontology, one of a set of ontologies intended to be used in pathway data annotation to ease data integration. This ontology is used to annotate protein names, protein family names, and generic/concrete protein names in the INOH pathway data. INOH is part of the BioPAX working group. |
Event (INOH pathway ontology) |
A structured controlled vocabulary of pathway-centric biological processes. This ontology is a INOH pathway annotation ontology, one of a set of ontologies intended to be used in pathway data annotation to ease data integration. This ontology is used to annotate biological processes, pathways, and sub-pathways in the INOH pathway data. INOH is part of the BioPAX working group. |
Protein-protein interaction |
A structured controlled vocabulary for the annotation of experiments concerned with protein-protein interactions. |
Pathway Ontology |
The Pathway Ontology is a controlled vocabulary for pathways that provides standard terms for the annotation of gene products. |
PATO |
PATO is an ontology of phenotypic qualities, intended for use in a number of applications, primarily phenotype annotation. For more information, please visit the PATO wiki (http://www.bioontology.org/wiki/index.php/PATO:Main_Page). |
Mouse pathology |
The Mouse Pathology Ontology (MPATH) is an ontology for mutant mouse pathology. This is Version 1. |
Human disease |
This ontology is a comprehensive hierarchical controlled vocabulary for human disease representation. For more information, please visit the Disease Ontology website (http://diseaseontology.sourceforge.net/). |
Although Cytoscape can import all kinds of ontologies in OBO format, annotation files are associated with specific ontologies. Therefore, you need to provide the correct ontology-specific annotation file to annotate nodes/edges/networks in Cytoscape. For example, while you can annotate human network data using the GO Full ontology with human Gene Association files, you cannot use a combination of the human Disease Ontology file and human Gene Association files, because the Gene Association file is only compatible with GO.
Visualize and Browse Ontology DAG (for Advanced Users)
Relationships between ontology terms are usually represented as Directed Acyclic Graphs (DAGs). This is a special case of a network (or graph), where nodes are ontology terms and edges are relationships between terms. Ontology data is stored in the same data structure as normal networks. This enables users and App writers to visualize, browse and manipulate ontology DAGs just like other networks. The following is an example of visualization of an ontology DAG (Generic GO Slim):
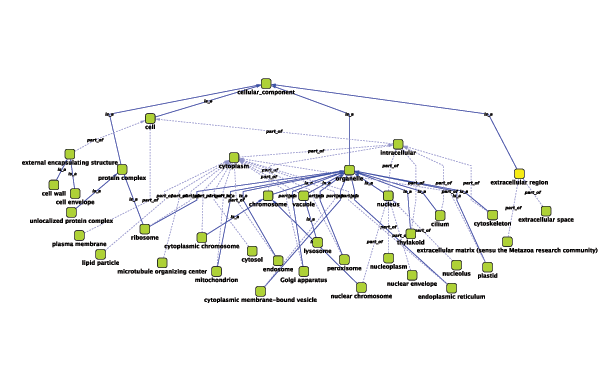
Every ontology term and relationship can have attributes. In the OBO files, ontology terms have optional fields such as definition, synonyms, comments, or cross-references. These fields will be imported as node attributes. To browse those attributes, please use the attribute browser (see the example below):
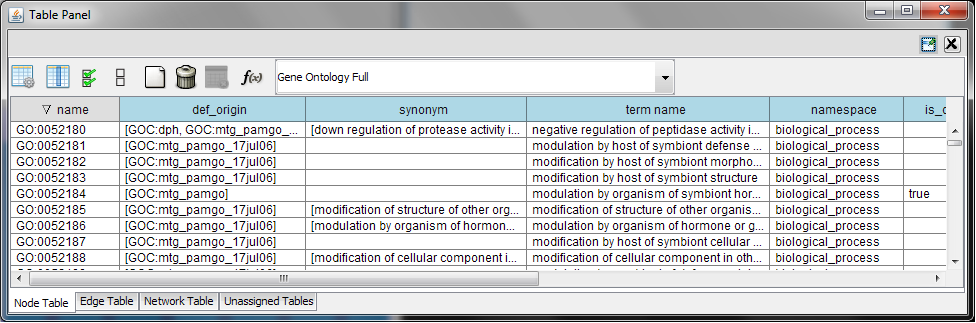
- Note 1: Some ontologies have a lot of terms. For example, the full Gene Ontology contains more than 20,000 terms. If you need to save memory, you can remove this ontology DAG from Network Panel (right-click on the ontology name at the left-hand side of the screen and select Destroy Network).
- Note 2: All ontology DAGs will be saved in the session file. To minimize the session file size, you can delete the Ontology DAG before saving session.
Gene Association File
The Gene Association (GA) file provides annotation only for the Gene Ontology. It is a species-specific annotation file for GO terms. Gene Association files will only work with Gene Ontologies and NOT others!
Sample Gene Association File (gene_association.sgd - annotation file for yeast):
SGD S000003916 AAD10 GO:0006081 SGD_REF:S000042151|PMID:10572264 ISS P aryl-alcohol dehydrogenase (putative) YJR155W gene taxon:4932 20020902 SGD SGD S000005275 AAD14 GO:0008372 SGD_REF:S000069584 ND C aryl-alcohol dehydrogenase (putative) YNL331C gene taxon:4932 20010119 SGD
Node Name Mapping
If you have a network file and an attribute file, they should have a common key to map attributes onto network data. If those two do not have a common key, you need to do an extra step to add a shared key. The following is a quick tutorial to learn how to use Gene Name Mapping files.
Import New ID Sets from BioMart
You can import various kinds of ID sets from BioMart (http://www.biomart.org/index.html). BioMart web service client is integrated into Cytoscape.
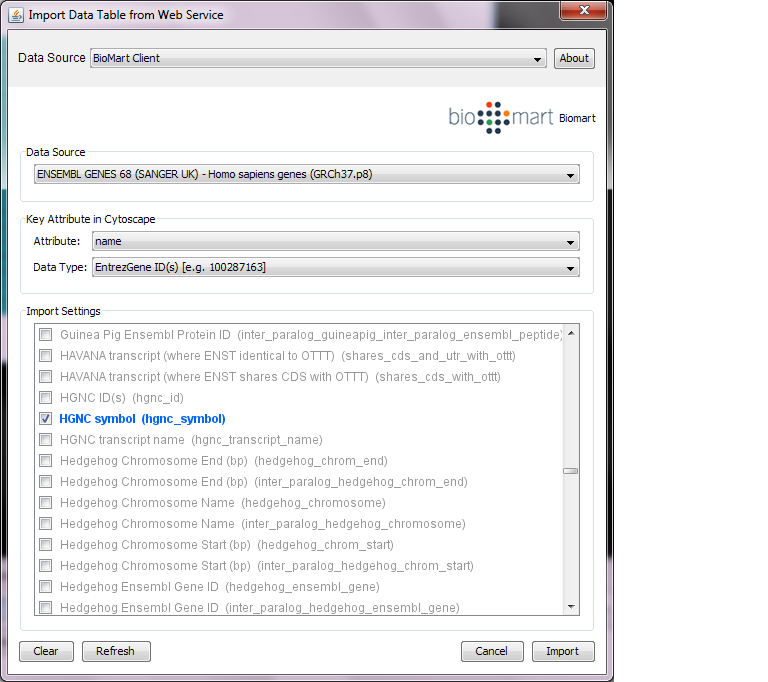
Select: File → Import → Table → Public Databases...
Select a data source. For ID mapping, select one of the Ensemble Genes data set. You need to choose correct species for your network.
Select Attribute. If you want to import new ID sets matching current node IDs, select name.
Select Data Type. This should be the type of ID set selected in Attribute list. For example, if you select name for Attribute and your network uses Entrez Gene ID for its node ID, you need to select EntrezGene ID(s) for Data Type.
Select new ID sets from the list. Because BioMart server does not accept query to import lots of annotations at once, you can select only 3-5 attributes for each import.
Press Import.
Import Gene Ontology and Gene Association Files
For convenience, Cytoscape has a list of URLs for commonly used ontology data and a complete set of Gene Association files. To import Gene Ontology and Gene Association files for the loaded networks, please follow these steps:
Important: All data sources in the preset list are remote URLs, meaning a network connection is required!
Step 1. Select an Annotation File
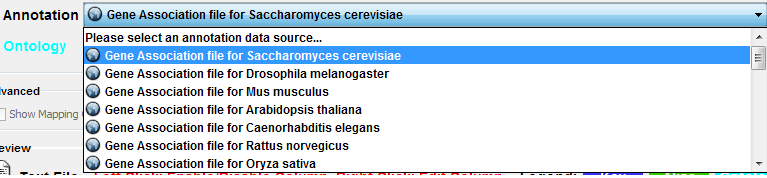
Select File → Import → Ontology and Annotation... to open the "Import Ontology and Annotation" window. From the Annotation dropdown list, select a gene association file for your network. For example, if you want to annotate the yeast network, select "Gene Association file for Saccharomyces cerevisiae".
Step 2. Select an Ontology File
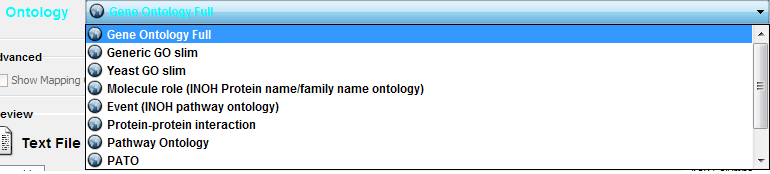
- Select an Ontology data (OBO file) from the Ontology dropdown list. If the file is not loaded yet, it will be shown in red. The first three files are Gene Ontology files. You can load other ontologies, but you need your own annotation file to annotate networks.
Step 3. Import the files
- Once you click the Import button, Cytoscape will start loading OBO and Gene Association files from the remote sources. If you choose GO Full it may take a while since it is a large data file.