Annotations in Cytoscape are stored as a set of ontologies (e.g. the Gene Ontology, or GO). An ontology consists of a set of controlled vocabulary terms that annotate the objects. For example, using the Gene Ontology, the Saccharomyces Cerevisiae CDC55 gene has a biological process described as “protein biosynthesis”, to which GO has assigned the number 6412 (a GO ID).
GO 8150 biological_process
- GO 7582 physiological processes
- GO 8152 metabolism
- GO 44238 primary metabolism
- GO 19538 protein metabolism
- GO 6412 protein biosynthesis
- GO 19538 protein metabolism
- GO 44238 primary metabolism
- GO 8152 metabolism
Graphical View of GO Term 6412: protein biosynthesis
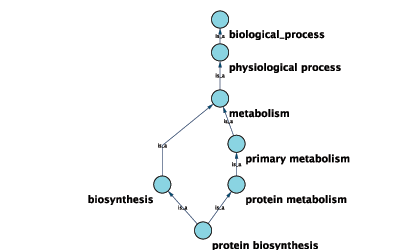
Cytoscape can use this ontology DAG (Directed Acyclic Graph) to annotate objects in networks. The Ontology Server (originally called "BioDataServer") is a Cytoscape feature which allows you to load, navigate, and assign annotation terms to nodes and edges in a network. Cytoscape 2.4 now has an enhanced GUI for loading ontology and associated annotation, enabling you to load both local and remote files.
Ontology and Annotation File Format
The standard file formats used in the Cytoscape Ontology Server are OBO and Gene Association. The GO website details these file formats:
Ontologies and Definitions: http://www.geneontology.org/GO.downloads.shtml#ont
Current Annotations: http://www.geneontology.org/GO.current.annotations.shtml