This is a legacy document
This page has been deprecated and is no longer updated. The current version of the Cytoscape manual can be found at http://manual.cytoscape.org/
GO 8150 biological_process
- GO 7582 physiological processes
- GO 8152 metabolism
- GO 44238 primary metabolism
- GO 19538 protein metabolism
- GO 6412 protein biosynthesis
- GO 19538 protein metabolism
- GO 44238 primary metabolism
- GO 8152 metabolism
Graphical View of GO Term 6412: protein biosynthesis
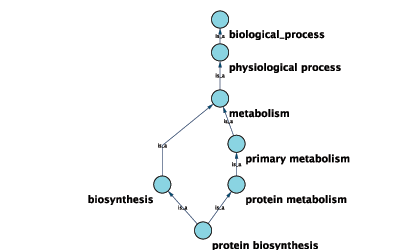
Cytoscape can use this ontology DAG (Directed Acyclic Graph) to annotate objects in networks. The Ontology Server (originally called "BioDataServer") is a Cytoscape feature which allows you to load, navigate, and assign annotation terms to nodes and edges in a network. Cytoscape 2.4 now has an enhanced GUI for loading ontology and associated annotation, enabling you to load both local and remote files.
Ontology and Annotation File Format
The standard file formats used in the Cytoscape Ontology Server are OBO and Gene Association. The GO website details these file formats:
Ontologies and Definitions: http://www.geneontology.org/GO.downloads.shtml#ont
Current Annotations: http://www.geneontology.org/GO.current.annotations.shtml
Default List of Ontologies
Cytoscape provides a list of ontologies available in OBO format. If an Internet connection is available, Cytoscape will import ontology and annotation files directly from the remote source. The table below lists the included ontologies.
Ontology Name |
Description |
Gene Ontology Full |
This data source contains a full-size GO DAG, which contains all GO terms. This OBO file is written in version 1.2 format. |
Generic GO slim |
A subset of general GO Terms, including higer-level terms only. |
Yeast GO slim |
A subset of GO Terms for annotating Yeast data sets maintained by SGD. |
Molecule role (INOH Protein name/family name ontology) |
A structured controlled vocabulary of concrete and abstract (generic) protein names. This ontology is a INOH pathway annotation ontology, one of a set of ontologies intended to be used in pathway data annotation to ease data integration. This ontology is used to annotate protein names, protein family names, and generic/concrete protein names in the INOH pathway data. INOH is part of the BioPAX working group. |
Event (INOH pathway ontology) |
A structured controlled vocabulary of pathway-centric biological processes. This ontology is a INOH pathway annotation ontology, one of a set of ontologies intended to be used in pathway data annotation to ease data integration. This ontology is used to annotate biological processes, pathways, and sub-pathways in the INOH pathway data. INOH is part of the BioPAX working group. |
Protein-protein interaction |
A structured controlled vocabulary for the annotation of experiments concerned with protein-protein interactions. |
Pathway Ontology |
The Pathway Ontology is a controlled vocabulary for pathways that provides standard terms for the annotation of gene products. |
PATO |
PATO is an ontology of phenotypic qualities, intended for use in a number of applications, primarily phenotype annotation. For more information, please visit the PATO wiki (http://www.bioontology.org/wiki/index.php/PATO:Main_Page). |
Mouse pathology |
The Mouse Pathology Ontology (MPATH) is an ontology for mutant mouse pathology. This is Version 1. |
Human disease |
This ontology is a comprehensive hierarchical controlled vocabulary for human disease representation. For more information, please visit the Disease Ontology website (http://diseaseontology.sourceforge.net/). |
Although Cytoscape can import all kinds of ontologies in OBO format, annotation files are associated with specific ontologies. Therefore, you need to provide the correct ontology-specific annotation file to annotate nodes/edges/networks in Cytoscape. For example, while you can annotate human network data using the GO Full ontology with human Gene Association files, you cannot use a combination of the human Disease Ontology file and human Gene Association files, because the Gene Association file is only compatible with GO.
Gene Association File
The Gene Association files provide annotation only for the Gene Ontology. It is a species-specific annotation file for GO terms. Gene Association files will only work with Gene Ontology annotation.
Sample Gene Association File (gene_association.sgd - annotation file for yeast):
SGD |
S000003916 |
AAD10 |
GO:0006081 |
SGD_REF:S000042151|PMID:10572264 |
ISS |
P |
aryl-alcohol dehydrogenase (putative) |
YJR155W gene |
taxon:4932 |
SGD |
S000005275 |
AAD14 |
GO:0008372 |
SGD_REF:S000069584 |
ND |
C |
aryl-alcohol dehydrogenase (putative) |
YNL331C gene |
taxon:4932 |
Import Ontology and Annotation
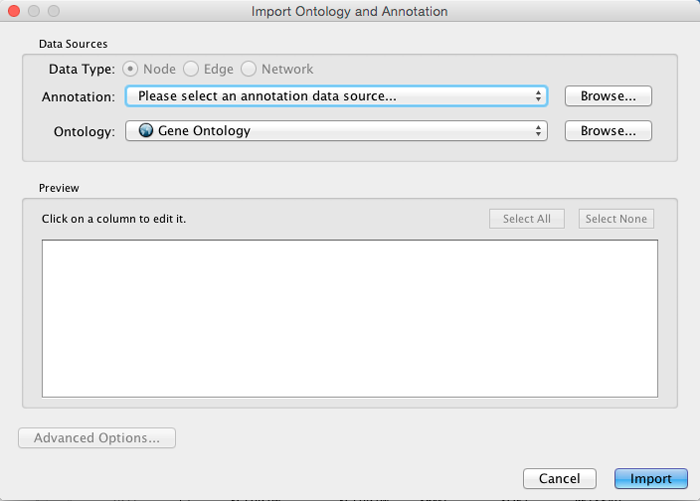
Cytoscape provides a graphical user interface to import both ontology and annotation files at the same time.
Note: All data sources in the preset list are remote URLs, meaning a network connection is required.
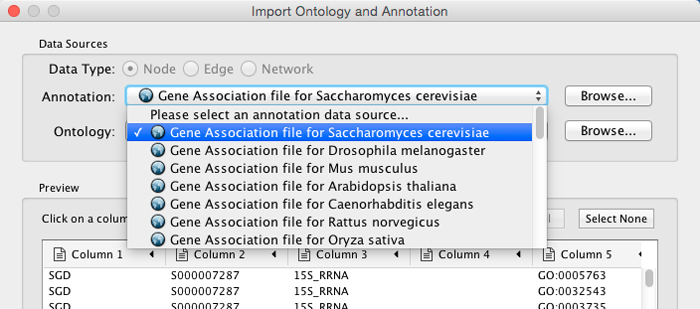
Select File → Import → Ontology and Annotation... to open the "Import Ontology and Annotation" interface. From the Annotation drop-down list, select a gene association file for your network. For example, if you want to annotate the yeast network, select "Gene Association file for Saccharomyces cerevisiae".
* Select an Ontology data (OBO file) from the Ontology drop-down list. If the file is not loaded yet, it will be shown in red. The first three files are Gene Ontology files. You can load other ontologies, but you need your own annotation file to annotate networks.
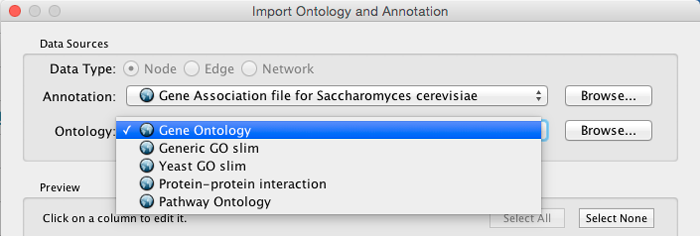
Once you click the Import button, Cytoscape will start loading OBO and Gene Association files from the remote sources. If you choose GO Full it may take a while since it is a large data file.
- When Cytoscape finishes importing files, the import window will be automatically closed. All columns mapped by this function have the prefix "annotation" and look like this: annotation.[column_name].
Note: Cytoscape supports both OBO formats: version 1.0 and 1.2.