RFC Name : CyNode identifier |
Editor(s): Ben Gross |
<<TableOfContents: execution failed [Argument "maxdepth" must be an integer value, not "[2]"] (see also the log)>>
About this document
This is an official Request for Comment (RFC) for CyNode Identification
For details on RFCs in general, check out the Wikipedia Entry: Request for Comments (RFCs)
Status
This RFC is still under construction and open for public comment. (01/17/06 -Ben)
How to Comment
To view/add comments, click on any of 'Comment' links below. By adding your ideas to the Wiki directly, we can more easily organize everyone's ideas, and keep clear records. Be sure to include today's date and your name for each comment. Here is an example to get things started: /Comment.
Try to keep your comments as concrete and constructive as possible. For example, if you find a part of the RFC makes no sense, please say so, but don't stop there. Take the extra step and propose alternatives.
Proposal
- Split Cytoscape.getCyNode(string alias, boolean create) into createCyNode(..) and getCyNode(string uid)
Any method that takes a string id to identify a CyNode should instead take a CyNode
Remove CANONICAL_NAME and COMMON_NAME /Comment
- Only use graph objects, not node indices, as parameters to methods, i.e. int [] getAdjacentEdges (Node node);
Still under discussion:
- There should be a clear distinction between a node id and its label.
Biological Questions / Use Cases
- User manipulates a node that has not been labeled e.g. when first created in the editor. (We need this now in the graph editor)
User wants to map nodes defined using UniProt IDs to nodes defined using Affymetrix IDs. (requires separation of externally defined node IDs from the unique CyNode IDs)
A use case previously tied to this issue, but is actually a NodeView issue:
- User views two or more nodes with the same unique identifier. E.g. ATP is highly connected in a metabolic network, so you may want to split it into two nodes in the view. (not yet implemented, but desired) Q: should the node also be split in the model?
General Notes
In this RFC, the term "label" or "node label" has been used in place of the more historic term "name" or "node name".
Requirements
Cytoscape need to have:
- nodes with a unique identifier (ID) that is unique for a session. It does not have to be globally unique or persist across sessions.
a standard way of attaching a string label to a CyNode that can be used to store e.g. the gene name
- the label and the unique identifier be separated.
- ability to import from and export to SIF, GML, node/edge attributes, gene expression data files.
Analysis
Cytoscape subsystems and their node identifier semantics
CyNetwork: Node ID in CyNode is a unique string (a GINY root graph index unique integer is also maintained and can be accessed by users). Semantics: the unique string is often expected to encode a gene name. Problems: two unique IDs are maintained (confusing for the developer), the unique string is used for gene names, which are not unique.
CyAttributes: A MultiHashMap using a unique string a key
SIF: Node ID is a unique string, often expected to encode a gene name. This is mapped to the CyNode unique string ID. Problem: user can define what the unique CyNode identifier is, thus the CyNode ID assumes different semantics depending on the user, which is a problem for graph merging.
GML: Node ID is a unique integer. A label attribute is available per node that is a string (does not have to be unique). This string label is mapped to CyNode unique string ID. Problem: a non-unique label is mapped to a unique string leading to potential conflicts.
Node attributes file format: Node ID is a unique string, often expected to encode a gene name. This is mapped to the CyNode unique string ID. Problem: user can define what the unique CyNode identifier is, thus the CyNode ID assumes different semantics depending on the user, which is a problem for matching node IDs in node attribute files to nodes defined in sif files.
Expression data file format: same semantics as SIF, but more often used to store expression IDs, like Affymetrix probeset IDs, while SIF often stored gene names. Problem: SIF and expression data IDs are often difficult to map to each other.
Attribute browser: currently displays CyNode string ID, canonical name, common name, aliases (these are often the same string - Problems: duplicated data, confusing for the user)
Merge plugin: merges based on CyNode unique string ID. Problem: this only works for networks where CyNode string IDs can be sensibly matched. If this is not the case, distinct nodes could be mistakenly merged. (TODO: need to check this with Ryan/Trey)
BioDataServer synonym table: Was formerly used for mapping IDs e.g. between SIF and expression data IDs, but is currently part of the code. Problem: synonym mapping is currently non-functional.
- Visual mapper: Uses nodeAttributes.getCanonicalName(node) to identify nodes.
Open Issues
SIF File Format:
- What new information (if any) has to be written to SIF ?
- SIF reader has to ensure that there are no duplicate labels.
- SIF reuses a label if it can find one.
- What effect does this have on new Cytoscape session saving subsystem ?
Edges:
- Should edges have an ID, or is a nodeID-edgeType-nodeID good enough to uniquely identify edges?
- Does graph merge depend on a unique string ID? Does it assume all identical nodes have the same root graph ID?
Backward Compatibility
Importing/Exporting:
- Ok to import old attributes file.
- Exporting should use new format.
Current Implementation Notes 2.2 (how a node gets created)
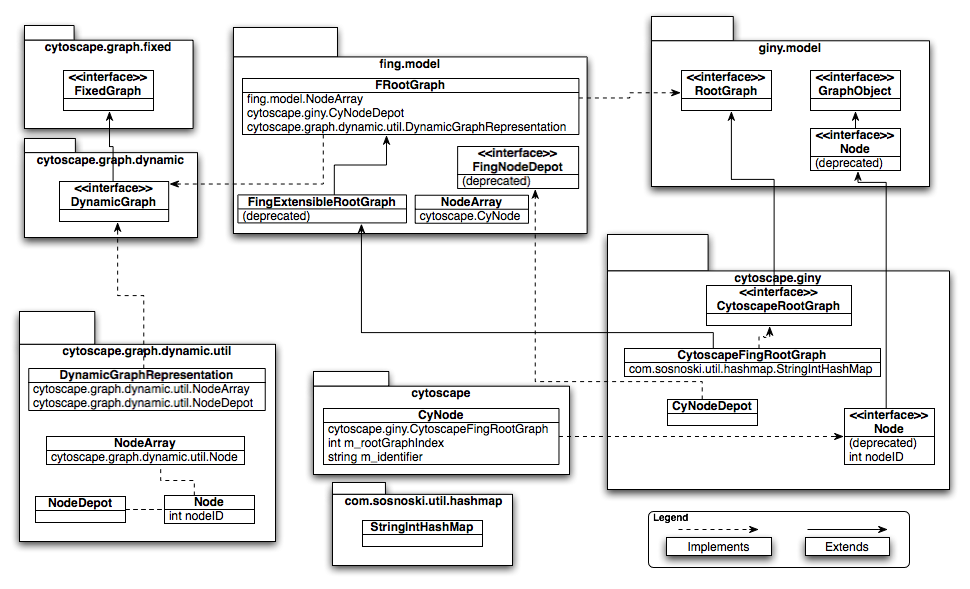
cytoscape.graph.dynamic.util.DynamicGraphRepresentation.nodeCreate() creates a unique node id as integer.
cytoscape.giny.CyNodeDepot creates new CyNode using nodeID obtained from cytoscape.graph.dynamic.util.DynamicGraphRepresentation.nodeCreate()
nodes are created/retrieved through Cytoscape.getCyNode(), using cytoscape.giny.CytoscapeFingRootGraph (which extends fing.model.FRootGraph). cytoscape.giny.CytoscapeFingRootGraph handles mapping of node indentifiers as strings to node id's as integers via com.sosnoski.util.hashmap.StringIntHashMap() (it translates the string to id before forwarding request to fing.model.FRootGraph).
- cytoscape.getCyNode() is called from:
- cytoscape.data.readers.GMLReader.read()
- cytoscape.data.readers.GMLReader2.createGraph()
cytoscape.data.readers.InteractionsReader.createRootGraphFromInteractionData()
cytoscape.editor.CytoscapeEditorManager.addNode()
Implementation Plan - Phase 1
Remove existing references to CANONICAL_NAME, COMMON_NAME, and ALIASES. This involved removing references to the Semantics class in the following classes:
- src/cytoscape/Cytoscape.java
- src/cytoscape/data/Semantics.java
src/cytoscape/data/readers/GMLReader2.java
src/cytoscape/data/readers/GMLTree.java
src/cytoscape/data/readers/VisualStyleBuilder.java
src/cytoscape/data/readers/XGMMLReader.java
src/cytoscape/data/writers/XGMMLWriter.java
src/cytoscape/dialogs/LabelTextPanel.java
src/cytoscape/layout/AttributeLayout.java
src/cytoscape/visual/CalculatorCatalog.java
In addition, an "ID" identifier was added to:
* src/cytoscape/visual/calculators/AbstractCalculator.java
to allow such things as a controlling attribute name to be set within a Mapping Class. In addition, the following classes were modified so that this new ID identifier would be added to the attribute bundle for each node and edge:
src/cytoscape/visual/calculators/GenericEdgeArrowCalculator.java
src/cytoscape/visual/calculators/GenericEdgeColorCalculator.java
src/cytoscape/visual/calculators/GenericEdgeFontFaceCalculator.java
src/cytoscape/visual/calculators/GenericEdgeFontSizeCalculator.java
src/cytoscape/visual/calculators/GenericEdgeLabelCalculator.java
src/cytoscape/visual/calculators/GenericEdgeLineTypeCalculator.java
src/cytoscape/visual/calculators/GenericEdgeToolTipCalculator.java
src/cytoscape/visual/calculators/GenericNodeColorCalculator.java
src/cytoscape/visual/calculators/GenericNodeFontFaceCalculator.java
src/cytoscape/visual/calculators/GenericNodeFontSizeCalculator.java
src/cytoscape/visual/calculators/GenericNodeLabelCalculator.java
src/cytoscape/visual/calculators/GenericNodeLabelColorCalculator.java
src/cytoscape/visual/calculators/GenericNodeLineTypeCalculator.java
src/cytoscape/visual/calculators/GenericNodeShapeCalculator.java
src/cytoscape/visual/calculators/GenericNodeSizeCalculator.java
src/cytoscape/visual/calculators/GenericNodeToolTipCalculator.java
Per Cytoscape conference calls, the vismapper code was going to be affected by this refactoring. It turns out the Gene Ontology/BioData Server - Gene Ontology Wizard is also affected by this refactoring. At least the following file makes references to canonical name:
src/cytoscape/data/annotation/AnnotationGui.java - no sure what this does
Implementation Plan - Phase 2
- Resolve open issues within RFC/clarify specification. Code analysis to support final RFC proposal.