Use Case Name : Network Clustering |
For Feature : Groups |
Editors: Gary Bader |
<<TableOfContents: execution failed [Argument "maxdepth" must be an integer value, not "[2]"] (see also the log)>>
Summary
A user would like to find densely connected regions in a network according to some criteria and view the region as a single node. This usually has the effect of simplifying the network and making higher order network structure more apparent.
Step-by-Step User Action
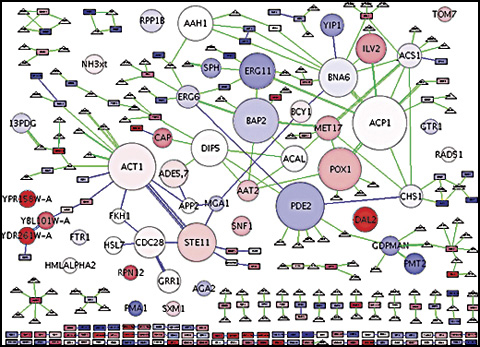
User runs network clustering algorithm, such as MCODE, BioModules or ActiveModules to detect 'modules' in the network. Modules are connected subnetworks satisfying some criteria, such as densely connected or transcriptionally active.
- User views results and chooses to collapse discovered modules into single nodes. The connections between the modules can be important.
Visual Aides
An example copied from http://galitski.systemsbiology.net/projs/biomodules/index.html
Requirements for Cytoscape
- Programmatic and user access to functions for grouping and ungrouping nodes. It is likely that all the above tools can use a common UI for allowing users to group/ungroup all modules or individual modules.
- Some algorithms may allow nodes to be included in multiple modules, so it would be nice if the grouping API supported that.
- Generally, it is fine to view just a simple circular node, not any fancy visualizations of the group.
Importance
- This is important for all network clustering algorithms, but doesn't preclude their use, so is not as important as some other use cases that are not currently possible.
Other Examples
- No known examples.