|
Size: 3047
Comment: Was having problems with one word Wiki names when used as sub pages
|
← Revision 7 as of 2009-02-12 01:03:30
Size: 3365
Comment: converted to 1.6 markup
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 9: | Line 9: |
|
|| '''Use Case Name''' : Degradation || || '''For Feature''' : Self:MIMEditor || |
|| '''Use Case Name''' : ../SpeciesDegradation || || '''For Feature''' : [[MIMEditor]] || |
| Line 18: | Line 18: |
| [[TableOfContents([2])]] | <<TableOfContents([2])>> |
| Line 23: | Line 23: |
| A user wants to describe the possibility that a protein is degraded as part of a StoichiometricConversion. | A user wants to describe the possibility that a protein is degraded as part of a ../StoichiometricConversion. |
| Line 29: | Line 29: |
| 1. User specifies a StoichiometricConversion with this protein | 1. User specifies a ../StoichiometricConversion with this protein |
| Line 38: | Line 38: |
| attachment:degradation1_061115_dwk.png | {{attachment:degradation1_061115_dwk.png}} |
| Line 42: | Line 42: |
| attachment:degradation2_061115_dwk.png | {{attachment:degradation2_061115_dwk.png}} |
| Line 63: | Line 63: |
| Shared MimEditorUseCaseComments | Shared ../MimEditorUseCaseComments |
| Line 65: | Line 65: |
| In the context of a StoichiometricConversion, the BioPax representation would basically be the same as any other StoichiometricConversion, except that the degradation products would be described as small molecules, which would not be used in subsequent reactions. | In the context of a ../StoichiometricConversion, the BioPax representation would basically be the same as any other ../StoichiometricConversion, except that the degradation products would be described as small molecules, which would not be used in subsequent reactions. |
| Line 67: | Line 67: |
| [[PageComment2]] |
---- AllanKuchinsky - 2007-01-22 05:15:47 Is there the need for special 'constant' Node(s) in Cytoscape to represent constructs like degradation? Are there special semantics associated with degradation 'nodes', e.g. would one ever want to assign attributes to a particular degradation 'node'? |
<<TableOfContents: execution failed [Argument "maxdepth" must be an integer value, not "[2]"] (see also the log)>>
Summary
A user wants to describe the possibility that a protein is degraded as part of a ../StoichiometricConversion.
Step-by-Step User Action
- User specifies a protein to be degraded
User specifies a ../StoichiometricConversion with this protein
- User specifies the result of the conversion is degradation
- User specifies the evidence for this relationship
Visual Aides
The Kohn notation for degradation is the following:

An example of the degradation symbol used as the result of a conversion follows:
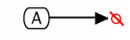
The color in this example is convention, and is not an essential element in the notation.
Requirements for Cytoscape
TBD
Importance
This is a fairly common reaction.
Other Examples
Comments
Shared ../MimEditorUseCaseComments
In the context of a ../StoichiometricConversion, the BioPax representation would basically be the same as any other ../StoichiometricConversion, except that the degradation products would be described as small molecules, which would not be used in subsequent reactions.
AllanKuchinsky - 2007-01-22 05:15:47
Is there the need for special 'constant' Node(s) in Cytoscape to represent constructs like degradation? Are there special semantics associated with degradation 'nodes', e.g. would one ever want to assign attributes to a particular degradation 'node'?