|
Size: 3526
Comment:
|
← Revision 9 as of 2009-02-12 01:03:45
Size: 5686
Comment: converted to 1.6 markup
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 1: | Line 1: |
|
## page was renamed from Molecular Interaction Maps/SimpleBinding ## page was renamed from SimpleBinding |
|
| Line 7: | Line 9: |
| || '''Use Case Name''' : SimpleBinding || | || '''Use Case Name''' : ../NonCovalentBinding || |
| Line 16: | Line 18: |
| [[TableOfContents([2])]] | <<TableOfContents([2])>> |
| Line 34: | Line 36: |
| attachment:simplebinding1a_061113_dwk.png | {{attachment:simplebinding1a_061113_dwk.png}} |
| Line 48: | Line 50: |
| == Comments == | == Variations == |
| Line 51: | Line 53: |
| attachment:simplebinding2a_061113_dwk.png | {{attachment:simplebinding2a_061113_dwk.png}} |
| Line 55: | Line 57: |
|
== Comments == Shared ../MimEditorUseCaseComments |
|
| Line 57: | Line 63: |
| The basic Kohn notation does not include a direct mechanism for attaching evidence to a particular reaction. In some of the eMims, we have added a text element A1, which can be subsequently linked to annotation information. | |
| Line 59: | Line 64: |
|
---- AllanKuchinsky - 2006-11-16 04:03:13 The right angle constraints will require 'snap-to-grid' functionality in the Cytoscape editor. This should be scheduled for implementation for Cytoscape 2.5. |
|
| Line 60: | Line 69: |
| [[PageComment2]] |
Would this also require a mechanism in the editor to *enforce* constraints such as Manhattan (right-angle) geometry under certain conditions? For example, when the user draws connecting edges for complexes in a MIMs-oriented editor, should the rubberbanding be restricted to horizontal or vertical lines? This should be worked into the editor framework for Cytoscape 2.5, perhaps via support in the API for a 'right-angle' flag for the BasicCytoShapeEntity class. But are there other, more general constraints that we need to support? That would require a more general constraint handling mechanism in the editor framework, which would require more thought. Hopefully, there is a finite set of constraints that we can handle directly, without the need for a general constraint handling mechanism. ---- GaryBader - 2006-11-16 07:40:25 This can also be represented as a 'physicalInteraction' in BioPAX as a shorthand. In this case, you do not have to specify that a complex is formed. However, this use is more general and you are not specifying the type of physical interaction. I.e. it could be a biochemical reaction, but the underlying experiment only tells you that there is a physical association. This is the kind of data generally stored in BIND, DIP, IntAct, MINT, HPRD, etc. ---- AllanKuchinsky - 2006-11-21 16:14:56 How does user specify evidence for an edge? Do we do this via contextual menu? By drag/drop? Would this be represented in Cytoscape via attribute(s)? Do we need a complex, structured 'evidence' attribute type, to handle the metadata as well as the data? ---- MiritAladjem - 2007-02-01 09:09:20 In response to Allan's last question: currently, evidence in the Kohn annotations is typically handled in the annotation file that is attached to the map. Interactions are numbered to correspoond to those annotations. This is an area where there is an opportunity to directly attach evidence to elements of the model. |
Use Case Name : ../NonCovalentBinding |
For Feature : MIM Editor |
Editors: DavidKane |
<<TableOfContents: execution failed [Argument "maxdepth" must be an integer value, not "[2]"] (see also the log)>>
Summary
A user wants to describe the possibility that two proteins can bind into a complex.
Step-by-Step User Action
- User specifies a protein to be involved in the interaction
- User specifies another protein to be involved in the interaction
- User specifies that there is a binding relationship between the two
- User specifies the evidence for this relationship
Visual Aides
The Kohn notation for this binding is the following:
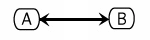
Requirements for Cytoscape
Best mapped to a group. A and B are connected and are part of the group. The edge might be a new node type. The visualization of the Kohn notation would require changes to the rendered and the vizmapper.
Importance
This is a basic building block of the Kohn notation.
Other Examples
Variations
A protein can be involved in more than one interaction. A protein can appear in a particular MIM once, and only once. If a protein is involved in additional reactions, then additional edges are added for each reaction.
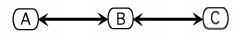
The layout rules for the edges for bindings require that they be horizontal or vertical, and may have any number of right angles. The edges may cross other edges at right angles.
Comments
Shared ../MimEditorUseCaseComments
The BioPAX representation of this binding could be a Conversion Object, i.e. with Participants = A,B, AB, Left = A,B, Right=AB, where A and B are Proteins, but AB is a complex with components A and B )
AllanKuchinsky - 2006-11-16 04:03:13
The right angle constraints will require 'snap-to-grid' functionality in the Cytoscape editor. This should be scheduled for implementation for Cytoscape 2.5.
Would this also require a mechanism in the editor to *enforce* constraints such as Manhattan (right-angle) geometry under certain conditions? For example, when the user draws connecting edges for complexes in a MIMs-oriented editor, should the rubberbanding be restricted to horizontal or vertical lines? This should be worked into the editor framework for Cytoscape 2.5, perhaps via support in the API for a 'right-angle' flag for the BasicCytoShapeEntity class. But are there other, more general constraints that we need to support? That would require a more general constraint handling mechanism in the editor framework, which would require more thought. Hopefully, there is a finite set of constraints that we can handle directly, without the need for a general constraint handling mechanism.
GaryBader - 2006-11-16 07:40:25
This can also be represented as a 'physicalInteraction' in BioPAX as a shorthand. In this case, you do not have to specify that a complex is formed. However, this use is more general and you are not specifying the type of physical interaction. I.e. it could be a biochemical reaction, but the underlying experiment only tells you that there is a physical association. This is the kind of data generally stored in BIND, DIP, IntAct, MINT, HPRD, etc.
AllanKuchinsky - 2006-11-21 16:14:56
How does user specify evidence for an edge? Do we do this via contextual menu? By drag/drop? Would this be represented in Cytoscape via attribute(s)? Do we need a complex, structured 'evidence' attribute type, to handle the metadata as well as the data?
MiritAladjem - 2007-02-01 09:09:20
In response to Allan's last question: currently, evidence in the Kohn annotations is typically handled in the annotation file that is attached to the map. Interactions are numbered to correspoond to those annotations. This is an area where there is an opportunity to directly attach evidence to elements of the model.