|
Size: 3458
Comment:
|
← Revision 8 as of 2009-02-12 01:04:04
Size: 3997
Comment: converted to 1.6 markup
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 4: | Line 4: |
|
## This template may be useful for documenting use cases ## Developed in response to a hackathon request for formalized, detailed use cases |
## This template may be useful for documenting use cases ## Developed in response to a hackathon request for formalized, detailed use cases |
| Line 12: | Line 12: |
| ## EXAMPLE: | ## EXAMPLE: |
| Line 17: | Line 17: |
| [[TableOfContents([2])]] | <<TableOfContents([2])>> |
| Line 35: | Line 35: |
| attachment:asymmetricbinding1_061115_dwk.png | {{attachment:asymmetricbinding1_061115_dwk.png}} |
| Line 39: | Line 39: |
| attachment:asymmetricbinding2_061115_dwk.png | {{attachment:asymmetricbinding2_061115_dwk.png}} |
| Line 53: | Line 53: |
| attachment:asymmetricbinding3_061115_dwk.png | {{attachment:asymmetricbinding3_061115_dwk.png}} |
| Line 60: | Line 60: |
| Shared ../MimEditorUseCaseComments | Shared ../MimEditorUseCaseComments |
| Line 63: | Line 63: |
| [[PageComment2]] |
* AllanKuchinsky - Does this mean that the dot notation is *only* used after a connection has been made to the reaction? This might require enhancements to the Vizmapper to support computed attributes -- or alternatively, enhancement to the editor to compute a hidden attribute on groups which would be used to conditionally render the dot. 2006-11-17 04:11:46 * AllanKuchinsky - Is there an implicit constraint on the edge between A and dot that the edge must *not* be of the type that would get the barbed edge in view? 2006-11-21 15:23:44 |
Use Case Name : ../AsymmetricBinding |
For Feature : MIMEditor |
Editors: DavidKane |
<<TableOfContents: execution failed [Argument "maxdepth" must be an integer value, not "[2]"] (see also the log)>>
Summary
A user wants to describe the possibility that a protein binds to another and contributes a peptide that binds to a receptor site or pocket on another protein.
Step-by-Step User Action
- User specifies a protein to be involved in the interaction as a contributor
- User specifies another protein to be involved in the reaction as a receptor
- User specifies that there is an asymmetric binding reaction between the two
- User optionally specifies the evidence for this reaction
Visual Aides
The Kohn notation for this reaction is the following:
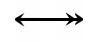
An example using this notation follows:
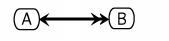
Requirements for Cytoscape
This would be modeled similarly to a ../NonCovalentBinding, i.e. with a group, but with a specific edge type. The barbed arrow would require custom rendering.
Importance
This notation is fairly rare and is not as important as the other use cases.
Variations
If the bound proteins are used in a subsequent reaction, a dot is placed on the line, and that is used to anchor the edges for the subsequent reactions. (See ../CompoundBinding)
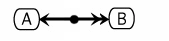
Other Examples
Comments
Shared ../MimEditorUseCaseComments
The BioPAX representation of this binding could be a Conversion Object, i.e. with Participants = A,B, AB, Left = A,B, Right=AB, where A and B are Proteins, but AB is a complex with components A and B). It is not clear how to differentiate simple binding from asymmetric binding in BioPAX.
AllanKuchinsky - Does this mean that the dot notation is *only* used after a connection has been made to the reaction? This might require enhancements to the Vizmapper to support computed attributes -- or alternatively, enhancement to the editor to compute a hidden attribute on groups which would be used to conditionally render the dot. 2006-11-17 04:11:46
AllanKuchinsky - Is there an implicit constraint on the edge between A and dot that the edge must *not* be of the type that would get the barbed edge in view? 2006-11-21 15:23:44