|
Size: 2777
Comment:
|
Size: 4087
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 37: | Line 37: |
|
==== Sample ==== * Network Overview {{attachment:sample_modules.png}} *Modules (A ~ D) {{attachment:moduleA.png}} {{attachment:moduleB.png}} {{attachment:moduleC.png}} {{attachment:moduleD.png}} |
|
| Line 41: | Line 54: |
| No extra information required. Users can use regular SIF file. |
We need to introduce two types of special value for '''''interaction''''' edge attributes: * '''''child_of''''' - Defines inclusion. * '''''interact_with''''' - interaction between modules. The sample graph above can be represented in the following: {{{ node7 DirectedEdge node3 node6 DirectedEdge node2 node5 DirectedEdge node4 node4 DirectedEdge node2 node4 DirectedEdge node3 node1 DirectedEdge node2 node1 DirectedEdge node0 node0 DirectedEdge node6 node0 DirectedEdge node2 moduleC child_of moduleA moduleD interact_with moduleA moduleD interact_with moduleB }}} If parser finds these two keywords in '''''interaction''''' edge attribute, treat it as a module relationship definition. |
| Line 47: | Line 81: |
|
subnet1 a b c subnet2 a d subnet3 c e f |
moduleA node0 node1 node2 node6 moduleB node3 node7 moduleC node0 node1 moduleD node2 node4 node5 |
| Line 51: | Line 86: |
|
Based on the information above, Cytoscape can rebuild the module relation map: {{attachment:moduleDAG.png}} Pros: simple and biologist-editable Cons: extra files needed |
|
| Line 55: | Line 97: |
|
==== Option 3: Simply Support DOT ==== As far as I know, DOT is the most complete solution for non-XML file format for representing modules. If we support this format, we can use it to represent |
Support for Modules in Cytoscape
Introduction
In many cases, biologists are interested in relationship between functional modules first. Then, they look into the details inside the modules, i.e., actual interactions. Cytoscape can partially handle this problem using Group API and some related plugins, but it's not a standard feature. Also, there is no simple file format to represent hierarchy/substructures. In this project, we are going to implement a mechanism to handle output of module finding algorithms as a hierarchy of subnetworks and make Cytoscape core module-aware.
Usecases
Module Finding Plugins
Currently, there are several plugins finding functional modules from large networks. In some cases, they produce subnetworks, but there is no universal UI or function to keep the structure in the session or other files.
Pathway Overview
Cytoscape has function to import KEGG/Reactome data as attributes. It is useful if Cytoscape has a function to generate substructure automatically based on such annotation.
Implementation
File Formats
Most of popular XML graph file formats support hierarchical structure.
DOT
This is a standard file format in graphviz. It has XML-like structure in the text file, and can represent substructure.
Pajek
This program has 3 types of files to represent substructure:
- Partitions – they tell for each vertex to which class vertex belong. Default extension: .clu.
- Clusters – subset of vertices (e.g. one class from partition). Default extension: .cls.
- Hierarchies – hierarchically ordered vertices. Default extension: .hie.
Therefore, user needs to load 4 files (network, partition, cluster, hierarchy) to reconstruct the saved substructures.
igraph
This package does not have its own file format to represents subgraph. Instead, supporting GraphML and dot.
Cytoscape New File Format for Subgraphs
Standard XML file formats such as GraphML or XGMML can represent substructures. However, they are not easy to edit by hand for many biologists. We need to implement a simple table/text style file format which is editable on spreadsheet programs.
Sample
- Network Overview
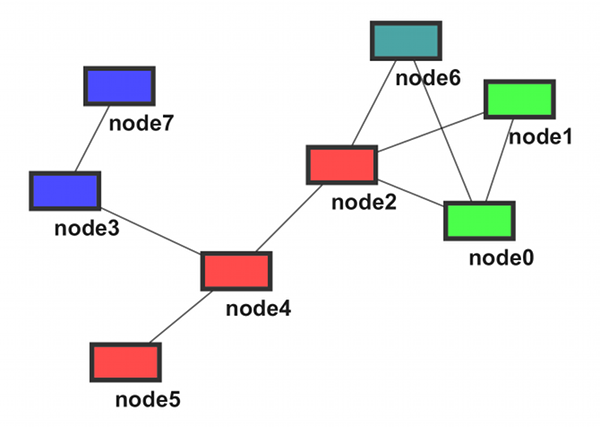
- Modules (A ~ D)
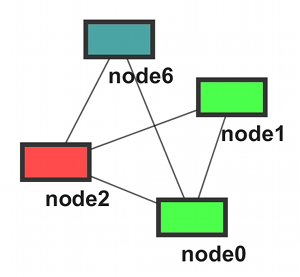
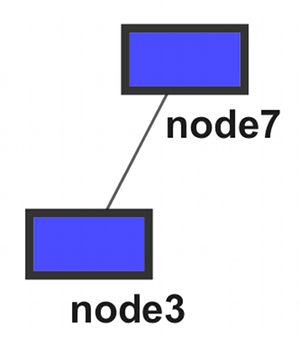
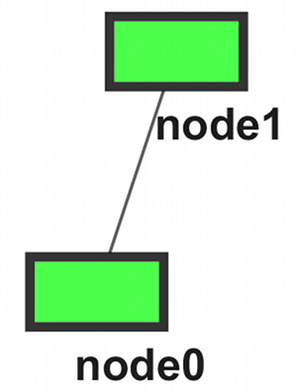
Option 1: Two Files
This approach needs two files, standard SIF file and subnetwork definition file (SDF?). This is similar to pajek, but less number of files are required to rebuild the substructures and hierarchies.
SIF
We need to introduce two types of special value for interaction edge attributes:
child_of - Defines inclusion.
interact_with - interaction between modules.
The sample graph above can be represented in the following:
node7 DirectedEdge node3 node6 DirectedEdge node2 node5 DirectedEdge node4 node4 DirectedEdge node2 node4 DirectedEdge node3 node1 DirectedEdge node2 node1 DirectedEdge node0 node0 DirectedEdge node6 node0 DirectedEdge node2 moduleC child_of moduleA moduleD interact_with moduleA moduleD interact_with moduleB
If parser finds these two keywords in interaction edge attribute, treat it as a module relationship definition.
SDF
This file defines members of each subnetwork.
moduleA node0 node1 node2 node6 moduleB node3 node7 moduleC node0 node1 moduleD node2 node4 node5
Based on the information above, Cytoscape can rebuild the module relation map:
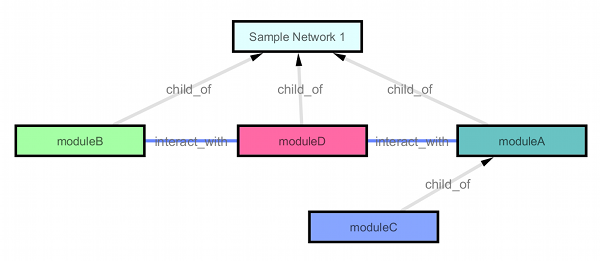
Pros: simple and biologist-editable Cons: extra files needed
Option 2: One File (extended SFI?)
(Not finished yet)
Option 3: Simply Support DOT
As far as I know, DOT is the most complete solution for non-XML file format for representing modules. If we support this format, we can use it to represent