How to Use Gene Name Mapping Files
This is a quick tutorial to learn how to use Gene Name Mapping files.
Import Network and Name Mapping Files
Download name mapping files. Mapping files are available at this page. In this tutorial, we are going to use dictionary_no_prefix.zip, which is a file set without prefixes for each gene names. Unzip the archive.
Load sample network file. Open network import dialog from File-->Import-->Network (multiple file types)... Then click URL radio button and import Human Protein-Protein: Rual et al. (Subnetwork for tutorial).
Open attribute table import dialog from File-->Import-->Attribute from Table.
Select human.dic_cyto.txt as the input file.
Check "Show Text File Import Options and click Transfer first line as attribute names checkbox.
Uncheck "Show Text File Import Options and check Mapping Options.
Select EntrezGene as Primary Key.
Right-click on EntrezGene column name and set the type to String.
Do the same for HGNC.
Right-click on Other Aliases and select List as the data type.
Check Other Aliases as Alias (under "Alias?" checkboxes).
- Now the Table Import dialog looks like the following screenshot:
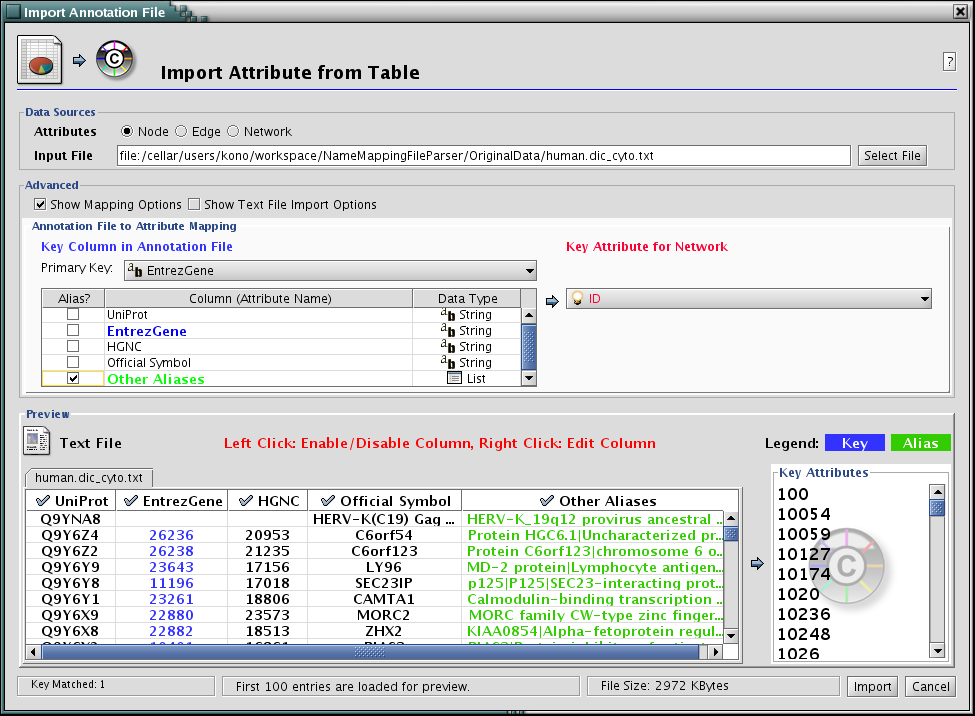
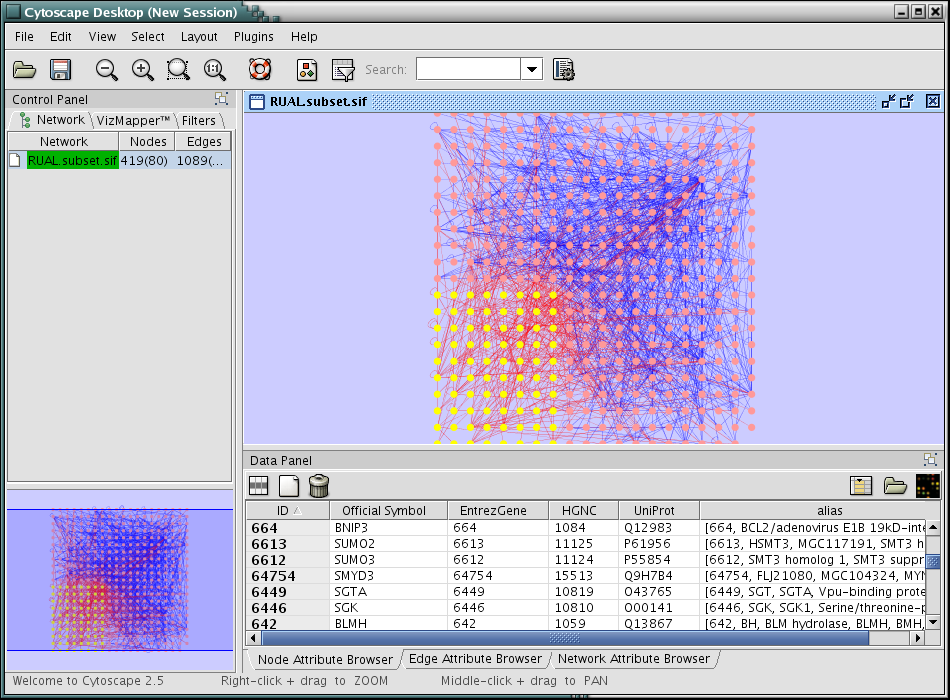
Press Import. The network has new names in the text file as attributes.
Annotate Network by Gene Association File
Note: the following tutorial is for Cytoscape 2.5 beta and later.
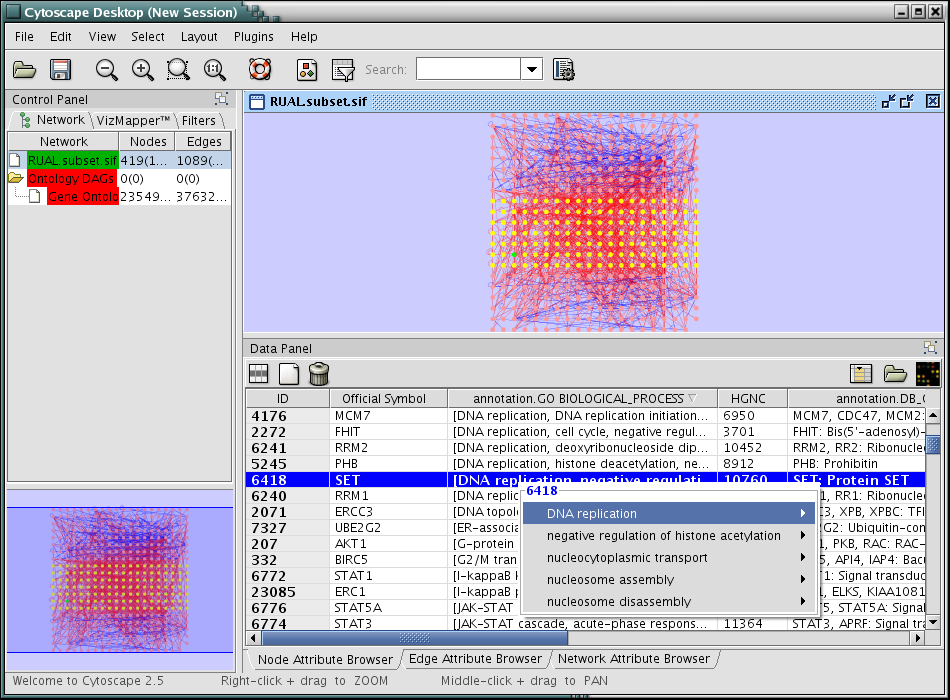
Open Gene Ontology import dialog: File-->Import-->Ontology and Annotation
From Annotation menu, select Gene Association file for Homo Sapiens.
Click Show Mapping Options.
Select DB_Object_ID as Primary Key.
Select UniProt for Key Attribute for Network.
- Press Import. The network is now annotated by Gene Association file!
Create Custom Mapping/Annotation Files by Ensembl Biomart
Ensembl Biomart is a web application to create custome annotation files from variety of data sources. You can create your own Cytoscape annotation files in few minutes by using biomart. The following is a tutorial to create custome annotation file for a human PPI network with EntrezGene ID as node identifier.
Load a human PPI network Human Protein-Protein: Rual et al. (Subnetwork for tutorial).
Go to Biomart web page.
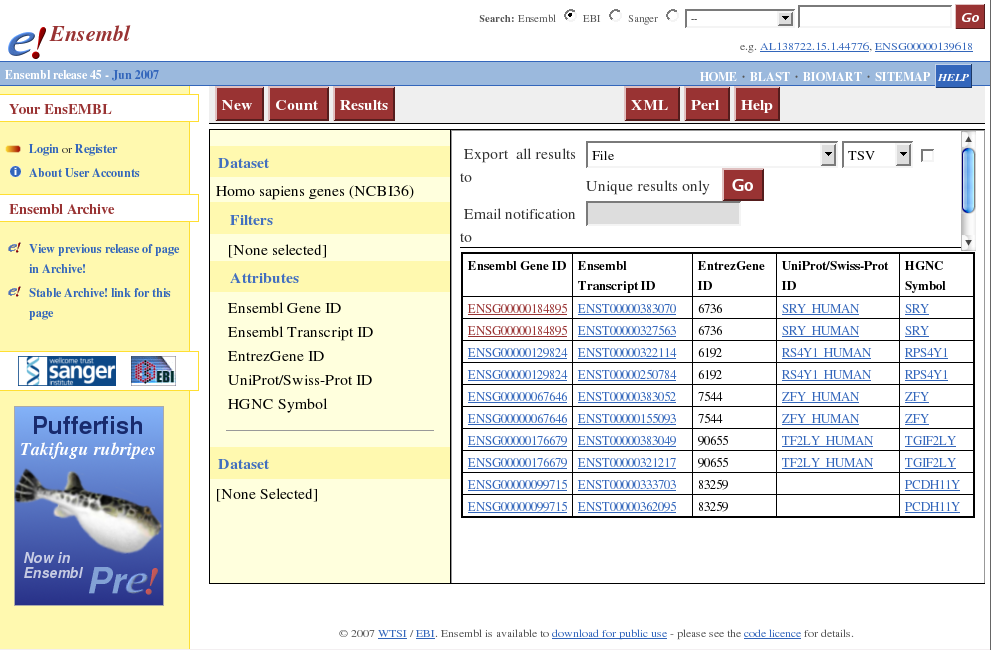
Select Ensembl 45 as data base, then select Homo sapiens genes (NCBI36) as dataset.
![[ATTACH] [ATTACH]](/moin_static192/modernized/img/attach.png)
Click Attributes. On the right, Features button is selected by default.
Click GENE. Ensembl Gene ID and Ensembl Transcript ID are selected. You can select as many attributes as you need.
Click EXTERNAL. Select EntrezGene ID. This will be used as the primary key for mapping. Select other attributes if necessary.
- Click other categories and select attributes you need to import.
Press Result button. Now you can see the result as HTML. (see the following)
Select Compressed file (.gz) from the combo box menu and make sure TSV (Tab-delimited text file) is selected.
Press GO. Biomart will create a compressed annotation file for you. Download process start automatically.
Go back to Cytoscape window and open attribute table import dialog from File-->Import-->Attribute from Table.
- Open the gzipped file from biomart. (Cytoscape supports compressed files, so you do not have to de-compress the file.)
Check Show Text File Import Options and click Transfer first line as attribute names checkbox.
Uncheck "Show Text File Import Options and check Mapping Options.
Select EntrezGene as Primary Key.
Right-click on EntrezGene column name and set the type to String.
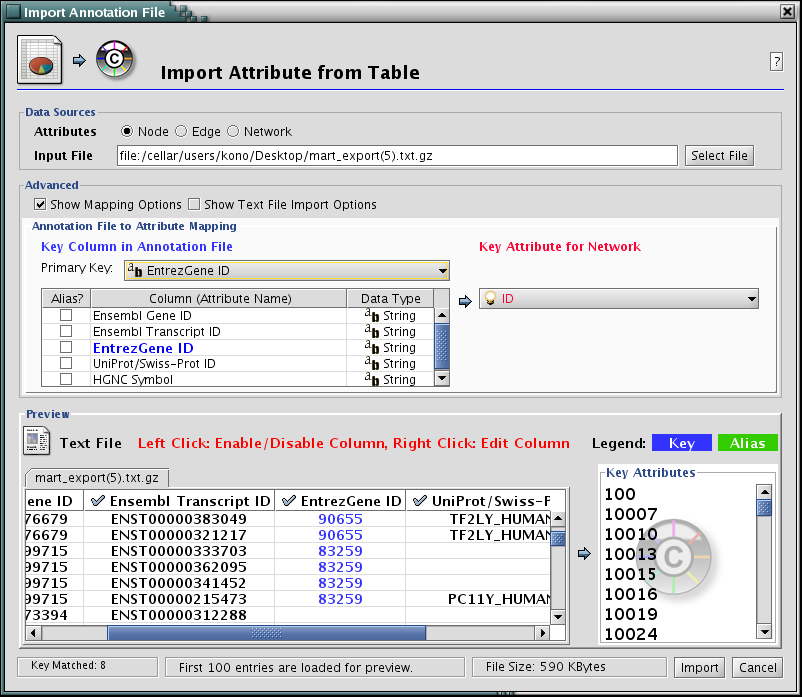
Press Import. New attributes and IDs are mapped onto the network.