|
Size: 10372
Comment:
|
Size: 19884
Comment: converted to 1.6 markup
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 1: | Line 1: |
| [[Anchor(navigation)]] | <<Anchor(navigation)>> |
| Line 4: | Line 4: |
| Cytoscape uses a [http://en.wikipedia.org/wiki/Zooming_User_Interface Zoomable User Interface] for navigating and viewing networks. ZUIs use two mechanisms for navigation: zooming and panning. Zooming increases or decreases the magnification of a view based on how much or how little a user wants to see. Panning allows users to move the focus of a screen to different parts of a view. | Cytoscape uses a [[http://en.wikipedia.org/wiki/Zooming_User_Interface|Zoomable User Interface]] for navigating and viewing networks. ZUIs use two mechanisms for navigation: zooming and panning. Zooming increases or decreases the magnification of a view based on how much or how little a user wants to see. Panning allows users to move the focus of a screen to different parts of a view. |
| Line 6: | Line 6: |
| '''Zoom''' Cytoscape provides two mechanisms for zooming, either using mouse gestures or buttons on the toolbar. Use the zooming buttons located on the toolbar to zoom in / out of the interaction network shown in the current network display. Zoom icons are detailed below: |
==== Zoom ==== Cytoscape provides two mechanisms for zooming: either using mouse gestures or buttons on the toolbar. Use the zooming buttons located on the toolbar to zoom in and out of the interaction network shown in the current network display. Zoom icons are detailed below: |
| Line 8: | Line 9: |
| attachment:image049.jpg | {{attachment:image049.jpg}} |
| Line 15: | Line 16: |
| * Zoom out to Display all of Current Network | * Zoom Out to Display all of Current Network |
| Line 19: | Line 20: |
| '''Pan''' You can pan the network image by holding down the middle mouse button and moving the mouse. You can also pan the image by holding down the left mouse button over the blue box in the Network Overview panel in the lower left hand of the Cytoscape desktop. |
==== Pan ==== You can pan the network image by holding down the middle mouse button and moving the mouse. You can also pan the image by holding down the left mouse button over the blue box in the Network Overview panel in the lower left hand of the Cytoscape desktop. |
| Line 22: | Line 24: |
| '''Select''' Click the left mouse button on a node or edge to select that object. You can hold down the Shift key to select more than one node/edge or you can hold down the left mouse button and drag the mouse to select groups of nodes/edges. |
==== Select ==== Click the left mouse button on a node or edge to select that object. You can hold down the Shift key to select more than one node/edge or you can hold down the left mouse button and drag the mouse to select groups of nodes/edges. |
| Line 24: | Line 27: |
| '''Context''' Click the right mouse button on a node/edge to launch a context sensitive menu with additional information about the node/edge. |
==== Context ==== Click the right mouse button on a node/edge to launch a context-sensitive menu with additional information about the node/edge. |
| Line 28: | Line 32: |
| The __Layout__ menu has an array of features for organizing the network visually according to one of several algorithms, aligning and rotating groups of nodes, and adjusting the size of the network. Most of these features are available from plugins that are packaged with Cytoscape 2.3. Some of the layout algorithms provided with Cytoscape 2.3 are: | The Layout menu has an array of features for organizing the network visually according to one of several algorithms, aligning and rotating groups of nodes, and adjusting the size of the network. Most of these features are available from plugins that are packaged with Cytoscape 2.3 and above. Cytoscape layouts have three different sources, which are reflected in the Layout menu. |
| Line 30: | Line 34: |
| '''Cytoscape Spring-embedded Layout ''' |
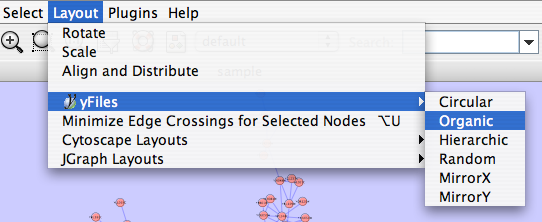
=== yFiles Layouts === '''yFiles''' layouts are a set of commercial layouts which are provided courtesy of [[http://www.yworks.com|yWorks]]. Due to license restrictions, the detailed parameters for these layouts are not available (there are no '''yFiles''' entries in the '''Settings...''' dialog). The main layout algorithms provided by yFiles are: |
| Line 32: | Line 37: |
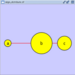
| Spring-embedded layout is based on a “force-directed” paradigm. Network nodes are treated like physical objects that repel each other, such as electrons. The connections between nodes are treated like metal springs attached to the pair of nodes. These springs repel or attract their end points according to a force function. The layout algorithm sets the positions of the nodes in a way that minimizes the sum of forces in the network. This algorithm is available from the __Layout → Cytoscape Layouts → Spring Embedded__ menu item. A sample screen shot is provided below: |
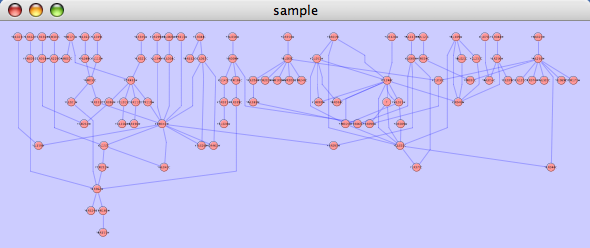
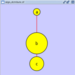
==== yFiles Circular Layout ==== This algorithm produces layouts that emphasize group and tree structures within a network. It partitions the network by analyzing its connectivity structure, and arranges the partitions as separate circles. The circles themselves are arranged in a radial tree layout fashion. This algorithm is available by selecting '''Layout → yFiles → Circular'''. {{attachment:image053.png}} |
| Line 34: | Line 41: |
|
attachment:image051.png '''yFiles Circular Layout ''' |
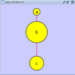
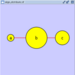
==== yFiles Hierarchical Layout ==== The hierarchical layout algorithm is good for representing main direction or "flow" within a network. Nodes are placed in hierarchically arranged layers and the ordering of the nodes within each layer is chosen in such a way that minimizes the number of edge crossings. This algorithm is available by selecting '''Layout → yFiles → Hierarchical'''. {{attachment:image055.png}} |
| Line 37: | Line 45: |
| This algorithm produces layouts that emphasize group and tree structures within a network. It partitions the network by analyzing its connectivity structure, and arranges the partitions as separate circles. The circles themselves are arranged in a radial tree layout fashion. This algorithm is available from the __Layout → yFiles → Circular__ menu item. |
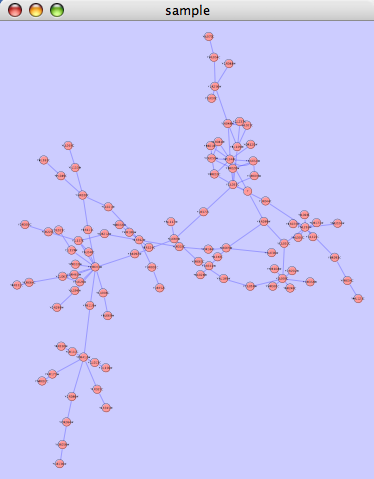
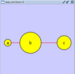
==== yFiles Organic Layout ==== The organic layout algorithm is a kind of spring-embedded algorithm that combines elements of the other algorithms to show the clustered structure of a graph. This algorithm is available by selecting '''Layout → yFiles → Organic'''. {{attachment:image057.png}} |
| Line 39: | Line 49: |
|
attachment:image053.png '''yFiles Hierarchical Layout ''' |
=== Cytoscape Layouts === |
| Line 42: | Line 51: |
| The hierarchical layout algorithm is good for representing main direction or ''flow'' within a network. Nodes are placed in hierarchically arranged layers and the ordering of the nodes within each layer is chosen in such a way that minimizes the number of edge crossings. This algorithm is available from the __Layout → yFiles → Heirarchical__ menu item. | '''Cytoscape Layouts''' are those layouts that have been written or integrated by Cytoscape developers. These layouts are fully integrated with Cytoscape. All Cytoscape Layouts have the option to operate on only the selected nodes, and all provide a '''Settings...''' panel to change the parameters of the algorithm. Most of the Cytoscape layouts also partition the graph before performing the layout. In addition, many of these layouts include the option to take either node or edge attributes into account. Some of these layouts are: |
| Line 44: | Line 53: |
|
attachment:image055.png '''yFiles Organic Layout ''' |
==== Grid Layout ==== The grid layout is a simple layout the arranges all of the nodes in a square grid. This is the default layout and is always available as part of the Cytoscape core. It is available by selecting '''Layout → Cytoscape Layouts → Grid Layout'''. A sample screen shot is shown below: {{attachment:grid_layout.png}} |
| Line 47: | Line 57: |
| The organic layout algorithm is a kind of spring-embedded algorithm that combines elements of the other algorithms to show the clustered structure of a graph. This algorithm is available from the __Layout → yFiles → Organic__ menu item. | ==== Force Directed Layout ==== |
| Line 49: | Line 59: |
| attachment:image057.png |
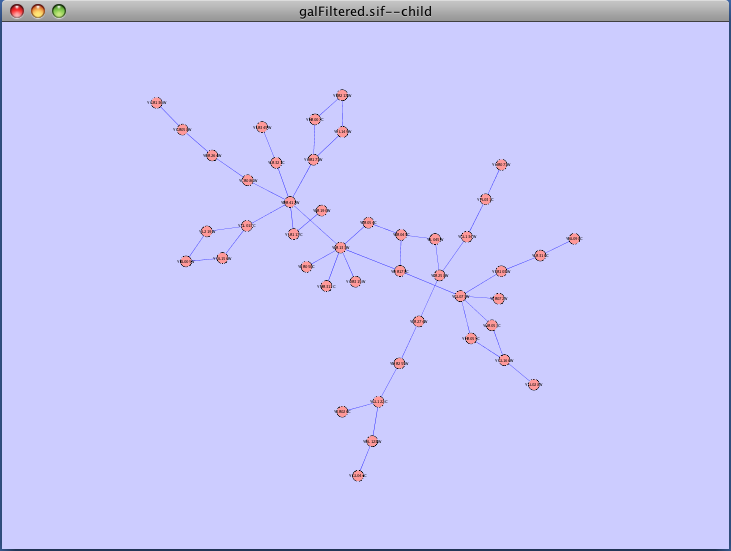
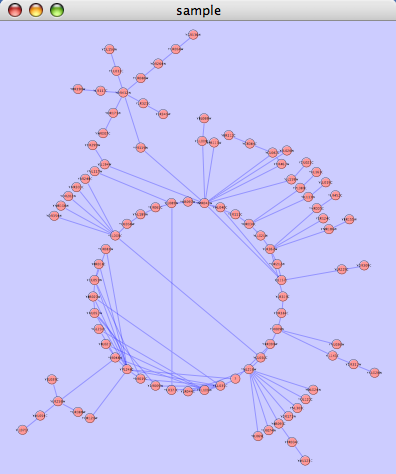
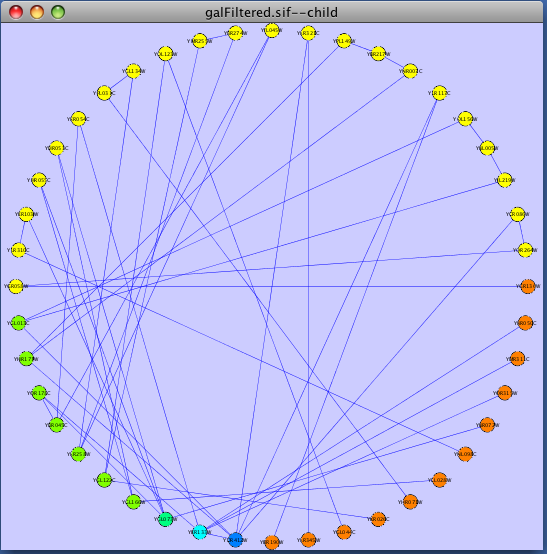
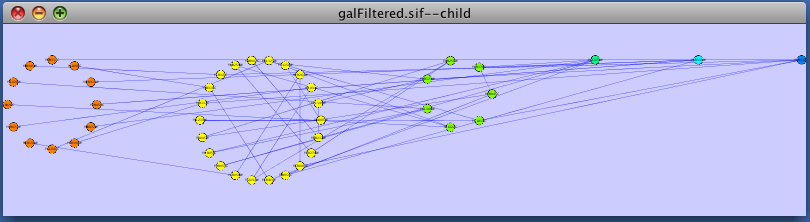
The force-directed layout is a layout based on the "force-directed" paradigm. This layout is based on the algorithm implemented as part of the excellent [[http://www.prefuse.org/|prefuse toolkit]] provided by Jeff Heer. The algorithm is very fast and with the right parameters can provide a very pleasing layout. The '''Force Directed Layout''' will also accept a numeric edge attribute to use as a weight for the length of the spring, although this will often require more use of the '''Settings...''' dialog to achieve the best layout. This algorithm is available by selecting '''Layout → Cytoscape Layouts → Force-Directed Layout → (unweighted)''' or the edge attribute you want to use as a weight. A sample screen shot showing a portion of the galFiltered network provided in sample data is provided below: {{attachment:force_layout.png}} ==== Spring-Embedded Layout ==== The spring-embedded layout is based on a “force-directed” paradigm as implemented by Kamada and Kawai (1988). Network nodes are treated like physical objects that repel each other, such as electrons. The connections between nodes are treated like metal springs attached to the pair of nodes. These springs repel or attract their end points according to a force function. The layout algorithm sets the positions of the nodes in a way that minimizes the sum of forces in the network. This algorithm can be applied to the entire network or a portion of it by selecting the appropriate options from '''Layout → Cytoscape Layouts → Spring Embedded'''. A sample screen shot is provided below: {{attachment:image051.png}} ==== Attribute Circle Layout ==== The attribute circle layout is a quick, useful layout, particularly for small networks, that will locate all of the nodes in the network around a circle. The node order is determined by a user-selected node attribute. The result is that all nodes with the same value for that attribute are located together around the circle. Using '''Layout → Cytoscape Layouts → Attribute Circle Layout →''' '''''attribute''''' to put all nodes around a circle using '''''attribute''''' to position them. The sample screen shot below shows the a subset of the galFiltered network organized by node Degree. {{attachment:attribute_circle_layout.png}} ==== Group Attributes Layout ==== The group attributes layout is similar to the attribute circle layout described above except that instead of a single circle with all of the nodes, each set of nodes that share the same value for the attribute are laid out in a separate circle. The same network shown above (subset of galFiltered) is shown below, using '''Layout → Cytoscape Layouts → Attribute Circle Layout → Degree'''. {{attachment:group_by_attributes_layout.png}} ==== Label Force-Directed Layout ==== The Label Force-Directed Layout is an automatic label repositioning plugin that can be accessed through '''Layout → Cytoscape Layouts → Label Force-Directed Layout'''. The layout is designed to be applied to position labels after another network layout is run to position nodes. It is compatible with any node positioning layout. Specific numerical values that the algorithm makes use of can be set by the user through the '''Layout → Settings...''' menu. Through adjusting these values, you can control how much you want to spread the labels and/or nodes of the network by. This label layout can be applied to the entire network, or only to those nodes that are selected (through enabling the ''Only layout selected nodes'' option). Upon the application of this plugin, node labels will be repositioned according to the Force-Directed algorithm, using the ''Default Label Spring Coefficient'' and ''Default Label Spring Length'' fields: * '''Default Label Spring Coefficient: ''' The strength of the pull between the label of a node and the node itself. The distance between nodes and their labels will be closer to the ''Default Label Spring Length'' if this value is larger. * '''Default Label Spring Length: ''' The desired length between the label and the node that the label belongs to. Users can also choose to move the network nodes. This option can be activated by enabling the ''Allow nodes to move'' option. All network nodes (or just the selected ones) will be moved according to the Force-Directed layout algorithm, while making use of the ''Default Spring Coefficient'', ''Default Spring Length'', and ''Default Percentage (%)'' fields: * '''Default Spring Coefficient: ''' The strength of the pull between a network node and its connected neighbours. The larger the value, the closer the distance between the two objects will be to the ''Default Spring Length'' value. * '''Default Spring Length: ''' The desired length of an edge that is connecting two network nodes together. * '''Default Percentage (%): ''' The percentage of the total ''Number of Iterations'' in which you would like the algorithm to move the network nodes as well, in addition to strictly moving the labels. For example: Number of Iterations = 200; Default Percentage (%) = 80. Then the network nodes will be moved for (80% / 100) * 200 = 160 of the 200 iterations. Other algorithm settings include: * '''Default Node Mass: ''' The mass that a default network node should take on. Nodes with a greater node mass require more strength to move. * '''Number of Iterations: ''' The total number of times to run the algorithm for each execution of this label layout. * '''Reset the label position of all nodes: ''' Reposition the label of each node to their default position. '''''Note: ''''' In order to yield the best layout for the labels, this plugin may need to be executed more than once. Before {{attachment:LabelForceDirectedLayout_Before.png}} After {{attachment:LabelForceDirectedLayout_After.png}} === JGraph Layouts === |
| Line 53: | Line 107: |
|
=== Layout Parameters === Many layouts have adjustable parameters that are exposed through the Layouts → Settings... menu option. This will pop up the following dialog, which allows you to choose which layout algorithm settings to adjust. The settings presented vary by algorithm and only those algorithms that allow access to their parameters will appear in the drop-down menu at the top of the dialog. Once you've modified a parameter, clicking the "Execute Layout" button will apply the layout. This be repeated until a useful layout is achieved. Settings can be saved using the "Save Settings" button. {{attachment:layout_settings_dialog.png}} |
|
| Line 54: | Line 116: |
| The simplest method to manually layout a network is to click on a node and drag the node. If you select multiple nodes, all of the selected nodes will be moved together. | The simplest method to manually organize a network is to click on a node and drag it. If you select multiple nodes, all of the selected nodes will be moved together. |
| Line 56: | Line 118: |
|
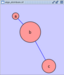
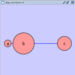
'''Edge Handles''' ''A little known feature!'' If you select an edge and then Ctrl-left-click on the edge, an edge "handle" will appear. This handle can be used to change the shape of the line. To remove a handle, simply Ctrl-left-click on the handle again. attachment:edge_handles.png The __Select → Edges__ menu has two menu items that provide further control: "Smooth Selected Edges" turns an edge consisting of line segments into a smoothed bezier curve, and "Straighen Selected Edges" turns a curved edge back into line segments. attachment:menu_select_edges.png '''Rotate''' attachment:rotate_dialog.png The __Layout → Rotate__ menu opens the Rotate dialog. Rotate will either rotate the entire network or a selected portion of the network. The image below shows a network with selected nodes rotated. |
==== Rotate ==== {{attachment:rotate_dialog.png}} Selecting the Layout → Rotate option will open the Rotate window in !CytoPanel 5. This function will either rotate the entire network or a selected portion of the network. The image below shows a network with selected nodes rotated. |
| Line 74: | Line 124: |
| attachment:rotate_networkA.png | {{attachment:rotate_networkA.png}} |
| Line 78: | Line 128: |
| attachment:rotate_networkB.png | {{attachment:rotate_networkB.png}} |
| Line 80: | Line 130: |
|
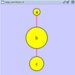
'''Scale''' attachment:scale_dialog.png The __Layout → Scale__ menu opens the Scale dialog. Scale will scale the position of the entire network or of the selected portion of the network. Note that only the position of the nodes scale, not the node sizes. Node size can be adjusted using the VizMapper. The image below shows selected nodes scaled. |
==== Scale ==== {{attachment:scale_dialog.png}} Selecting the Layout → Scale option will open the Scale window in !CytoPanel 5. This function will scale the position of the entire network or of the selected portion of the network. Note that only the position of the nodes will be scaled, not the node sizes. Node size can be adjusted using the !VizMapper. The image below shows selected nodes scaled. |
| Line 88: | Line 136: |
| attachment:scale_networkA.png | {{attachment:scale_networkA.png}} |
| Line 92: | Line 140: |
| attachment:scale_networkB.png | {{attachment:scale_networkB.png}} |
| Line 94: | Line 142: |
|
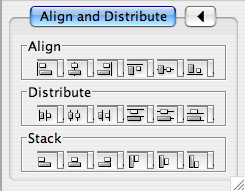
'''Align/Distribute''' attachment:align_distribute_dialog.png The __Layout → Align/Distribute__ menu opens the Align/Distribute dialog. The Align buttons of the dialog provides different options for either vertically or horizontally aligning selected nodes against a line. The differences are in what part of the node gets aligned, e.g. the center of the node, the top of the node, the left side of the node. The Distribute buttons evenly distribute selected nodes between the two most distant nodes along either the vertical or horizontal axis. The differences are again a function what part of the node is used as a reference point for the distribution. The table below provides a mapping from button to name to result. |
==== Align/Distribute/Stack ==== {{attachment:align_distribute_dialog.png}} Selecting the Layout → Align/Distribute option will open the Align/Distribute/Stack window in !CytoPanel 5. The Align buttons provide different options for either vertically or horizontally aligning selected nodes against a line. The differences are in what part of the node gets aligned, e.g. the center of the node, the top of the node, the left side of the node. The Distribute buttons evenly distribute selected nodes between the two most distant nodes along either the vertical or horizontal axis. The differences are again a function what part of the node is used as a reference point for the distribution. And the Stack buttons vertically or horizontally stack selected nodes with the full complement of alignment options. The table below provides a decription of what each button does. |
| Line 101: | Line 147: |
|
|| attachment:V_ALIGN_TOP.gif ||attachment:small.align_begin.png||attachment:small.align_vertical_top.png||Vertical Align Top - The tops of the selected nodes are aligned with the top-most node. || || attachment:V_ALIGN_CENTER.gif ||attachment:small.align_begin.png||attachment:small.align_vertical_center.png||Vertical Align Center - The centers of selected nodes are aligned along a line defined by the midpoint between the top and bottom-most nodes. || || attachment:V_ALIGN_BOTTOM.gif ||attachment:small.align_begin.png||attachment:small.align_vertical_bottom.png||Vertical Align Bottom - The bottoms of the nodes are aligned with the bottom-most node. || || attachment:H_ALIGN_LEFT.gif ||attachment:small.align_begin.png||attachment:small.align_horizontal_left.png||Horizontal Align Left - The left hand sides of the selected nodes are aligned with the left-most node. || || attachment:H_ALIGN_CENTER.gif ||attachment:small.align_begin.png||attachment:small.align_horizontal_center.png||Horizontal Align Center - The centers of selected nodes are aligned along a line defined by the midpoint between the left and right-most nodes. || || attachment:H_ALIGN_RIGHT.gif ||attachment:small.align_begin.png||attachment:small.align_horizontal_right.png||Horizontal Align Right - The right hand sides of the selected nodes are aligned with the right-most node. || |
|| {{attachment:V_ALIGN_TOP.gif}} ||{{attachment:small.align_begin.png}}||{{attachment:small.align_vertical_top.png}}||Vertical Align Top - The tops of the selected nodes are aligned with the top-most node. || || {{attachment:V_ALIGN_CENTER.gif}} ||{{attachment:small.align_begin.png}}||{{attachment:small.align_vertical_center.png}}||Vertical Align Center - The centers of the selected nodes are aligned along a line defined by the midpoint between the top and bottom-most nodes. || || {{attachment:V_ALIGN_BOTTOM.gif}} ||{{attachment:small.align_begin.png}}||{{attachment:small.align_vertical_bottom.png}}||Vertical Align Bottom - The bottoms of the selected nodes are aligned with the bottom-most node. || || {{attachment:H_ALIGN_LEFT.gif}} ||{{attachment:small.align_begin.png}}||{{attachment:small.align_horizontal_left.png}}||Horizontal Align Left - The left hand sides of the selected nodes are aligned with the left-most node. || || {{attachment:H_ALIGN_CENTER.gif}} ||{{attachment:small.align_begin.png}}||{{attachment:small.align_horizontal_center.png}}||Horizontal Align Center - The centers of the selected nodes are aligned along a line defined by the midpoint between the left and right-most nodes. || || {{attachment:H_ALIGN_RIGHT.gif}} ||{{attachment:small.align_begin.png}}||{{attachment:small.align_horizontal_right.png}}||Horizontal Align Right - The right hand sides of the selected nodes are aligned with the right-most node. || |
| Line 110: | Line 156: |
|
|| attachment:V_DIST_TOP.gif ||attachment:small.distribute_begin_vertical.png||attachment:small.distribute_vertical_top.png||Vertical Distribute Top - The tops of the selected nodes are distributed evenly between the top-most and bottom-most nodes, which should stay stationary. || || attachment:V_DIST_CENTER.gif ||attachment:small.distribute_begin_vertical.png||attachment:small.distribute_vertical_center.png||Vertical Distribute Center - The centers of the selected nodes are distributed evenly between the top-most and bottom-most nodes, which should stay stationary. || || attachment:V_DIST_BOTTOM.gif ||attachment:small.distribute_begin_vertical.png||attachment:small.distribute_vertical_bottom.png||Vertical Distribute Bottom - The bottoms of the selected nodes are distributed evenly between the top-most and bottom-most nodes, which should stay stationary. || || attachment:H_DIST_LEFT.gif ||attachment:small.distribute_begin_horizontal.png||attachment:small.distribute_horizontal_left.png||Horizontal Distribute Left - The left hand sides of the selected nodes are distributed evenly between the left-most and rigth-most nodes, which should stay stationary. || || attachment:H_DIST_CENTER.gif ||attachment:small.distribute_begin_horizontal.png||attachment:small.distribute_horizontal_center.png||Horizontal Distribute Center - The centers of the selected nodes are distributed evenly between the left-most and rigth-most nodes, which should stay stationary. || || attachment:H_DIST_RIGHT.gif ||attachment:small.distribute_begin_horizontal.png||attachment:small.distribute_horizontal_right.png||Horizontal Distribute Right - The right hand sides of the selected nodes are distributed evenly between the left-most and rigth-most nodes, which should stay stationary. || |
|| {{attachment:V_DIST_TOP.gif}} ||{{attachment:small.distribute_begin_vertical.png}}||{{attachment:small.distribute_vertical_top.png}}||Vertical Distribute Top - The tops of the selected nodes are distributed evenly between the top-most and bottom-most nodes, which should stay stationary. || || {{attachment:V_DIST_CENTER.gif}} ||{{attachment:small.distribute_begin_vertical.png}}||{{attachment:small.distribute_vertical_center.png}}||Vertical Distribute Center - The centers of the selected nodes are distributed evenly between the top-most and bottom-most nodes, which should stay stationary. || || {{attachment:V_DIST_BOTTOM.gif}} ||{{attachment:small.distribute_begin_vertical.png}}||{{attachment:small.distribute_vertical_bottom.png}}||Vertical Distribute Bottom - The bottoms of the selected nodes are distributed evenly between the top-most and bottom-most nodes, which should stay stationary. || || {{attachment:H_DIST_LEFT.gif}} ||{{attachment:small.distribute_begin_horizontal.png}}||{{attachment:small.distribute_horizontal_left.png}}||Horizontal Distribute Left - The left hand sides of the selected nodes are distributed evenly between the left-most and right-most nodes, which should stay stationary. || || {{attachment:H_DIST_CENTER.gif}} ||{{attachment:small.distribute_begin_horizontal.png}}||{{attachment:small.distribute_horizontal_center.png}}||Horizontal Distribute Center - The centers of the selected nodes are distributed evenly between the left-most and right-most nodes, which should stay stationary. || || {{attachment:H_DIST_RIGHT.gif}} ||{{attachment:small.distribute_begin_horizontal.png}}||{{attachment:small.distribute_horizontal_right.png}}||Horizontal Distribute Right - The right hand sides of the selected nodes are distributed evenly between the left-most and right-most nodes, which should stay stationary. || ||'''Button'''||'''Before'''||'''After'''||'''Description of Stack Options''' || || {{attachment:V_STACK_LEFT.gif}} ||{{attachment:small.stack_vertical_begin.png}}||{{attachment:small.stack_vertical_left.png}}||Vertical Stack Left - Vertically stacked below top-most node with the left-hand sides of the selected nodes aligned. || || {{attachment:V_STACK_CENTER.gif}} ||{{attachment:small.stack_vertical_begin.png}}||{{attachment:small.stack_vertical_center.png}}||Vertical Stack Center - Vertically stacked below top-most node with the centers of selected nodes aligned. || || {{attachment:V_STACK_RIGHT.gif}} ||{{attachment:small.stack_vertical_begin.png}}||{{attachment:small.stack_vertical_right.png}}||Vertical Stack Right - Vertically stacked below top-most node with the right-hand sides of the selected nodes aligned. || || {{attachment:H_STACK_TOP.gif}} ||{{attachment:small.stack_horizontal_begin.png}}||{{attachment:small.stack_horizontal_top.png}}||Horizontal Stack Top - Horizontally stacked to the right of the left-most node with the tops of the selected nodes aligned. || || {{attachment:H_STACK_CENTER.gif}} ||{{attachment:small.stack_horizontal_begin.png}}||{{attachment:small.stack_horizontal_center.png}}||Horizontal Stack Center - Horizontally stacked to the right of the left-most node with the centers of selected nodes aligned. || || {{attachment:H_STACK_BOTTOM.gif}} ||{{attachment:small.stack_horizontal_begin.png}}||{{attachment:small.stack_horizontal_bottom.png}}||Horizontal Stack Bottom - Horizontal Stack Center - Horizontally stacked to the right of the left-most node with the bottoms of the selected nodes aligned. || ==== Edge Handles ==== This is a little-known feature! If you select an edge and then Ctrl-left-click on the edge, an edge "handle" will appear. This handle can be used to change the shape of the line. To remove a handle, simply Ctrl-left-click on the handle again. ||Straight Edge ||Smooth Edge || ||{{attachment:edge_handles1.png}}||{{attachment:edge_handles2.png}}|| Two other options under Select → Edges provide further control: "Smooth Selected Edges" turns an edge consisting of line segments into a smoothed bezier curve, and "Straighten Selected Edges" turns a curved edge back into line segments. {{attachment:menu_select_edges.png}} |
| Line 119: | Line 184: |
| In addition to the ability to click on a node and drag it to a new position, Cytoscape now has the ability move nodes using the arrow keys on the keyboard. By selecting one or more nodes using the mouse and clicking one of the arrow keys (←, →, ↑, ↓) the selected nodes will move one pixel in the chosen direction. If an arrow key is pressed while holding the {{{shift}}} key down, the selected nodes will 10 pixels in the chosen direction. | In addition to the ability to click on a node and drag it to a new position, Cytoscape now has the ability to move nodes using the arrow keys on the keyboard. By selecting one or more nodes using the mouse and clicking one of the arrow keys (←, →, ↑, ↓) the selected nodes will move one pixel in the chosen direction. If an arrow key is pressed while holding the Shift key down, the selected nodes will 10 pixels in the chosen direction. |
| Line 121: | Line 186: |
| Mouse movement has also been enhanced. If the {{{shift}}} key is held down while dragging a node, the node will only move horizontally, vertically, or along a 45 degree diagonal. | Mouse movement has also been enhanced. If the Shift key is held down while dragging a node, the node will only move horizontally, vertically, or along a 45 degree diagonal. |
Basic Network Navigation
Cytoscape uses a Zoomable User Interface for navigating and viewing networks. ZUIs use two mechanisms for navigation: zooming and panning. Zooming increases or decreases the magnification of a view based on how much or how little a user wants to see. Panning allows users to move the focus of a screen to different parts of a view.
Zoom
Cytoscape provides two mechanisms for zooming: either using mouse gestures or buttons on the toolbar. Use the zooming buttons located on the toolbar to zoom in and out of the interaction network shown in the current network display. Zoom icons are detailed below:
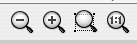
From Left to Right:
- Zoom Out
- Zoom In
- Zoom Selected Region
- Zoom Out to Display all of Current Network
You can also zoom in/out by holding down the right mouse button and moving the mouse to the right (zoom in) or left (zoom out).
Pan
You can pan the network image by holding down the middle mouse button and moving the mouse. You can also pan the image by holding down the left mouse button over the blue box in the Network Overview panel in the lower left hand of the Cytoscape desktop.
Other Mouse Behaviors
Select
Click the left mouse button on a node or edge to select that object. You can hold down the Shift key to select more than one node/edge or you can hold down the left mouse button and drag the mouse to select groups of nodes/edges.
Context
Click the right mouse button on a node/edge to launch a context-sensitive menu with additional information about the node/edge.
Automatic Layout Algorithms
The Layout menu has an array of features for organizing the network visually according to one of several algorithms, aligning and rotating groups of nodes, and adjusting the size of the network. Most of these features are available from plugins that are packaged with Cytoscape 2.3 and above. Cytoscape layouts have three different sources, which are reflected in the Layout menu.
yFiles Layouts
yFiles layouts are a set of commercial layouts which are provided courtesy of yWorks. Due to license restrictions, the detailed parameters for these layouts are not available (there are no yFiles entries in the Settings... dialog). The main layout algorithms provided by yFiles are:
yFiles Circular Layout
This algorithm produces layouts that emphasize group and tree structures within a network. It partitions the network by analyzing its connectivity structure, and arranges the partitions as separate circles. The circles themselves are arranged in a radial tree layout fashion. This algorithm is available by selecting Layout → yFiles → Circular.
yFiles Hierarchical Layout
The hierarchical layout algorithm is good for representing main direction or "flow" within a network. Nodes are placed in hierarchically arranged layers and the ordering of the nodes within each layer is chosen in such a way that minimizes the number of edge crossings. This algorithm is available by selecting Layout → yFiles → Hierarchical.
yFiles Organic Layout
The organic layout algorithm is a kind of spring-embedded algorithm that combines elements of the other algorithms to show the clustered structure of a graph. This algorithm is available by selecting Layout → yFiles → Organic.
Cytoscape Layouts
Cytoscape Layouts are those layouts that have been written or integrated by Cytoscape developers. These layouts are fully integrated with Cytoscape. All Cytoscape Layouts have the option to operate on only the selected nodes, and all provide a Settings... panel to change the parameters of the algorithm. Most of the Cytoscape layouts also partition the graph before performing the layout. In addition, many of these layouts include the option to take either node or edge attributes into account. Some of these layouts are:
Grid Layout
The grid layout is a simple layout the arranges all of the nodes in a square grid. This is the default layout and is always available as part of the Cytoscape core. It is available by selecting Layout → Cytoscape Layouts → Grid Layout. A sample screen shot is shown below:
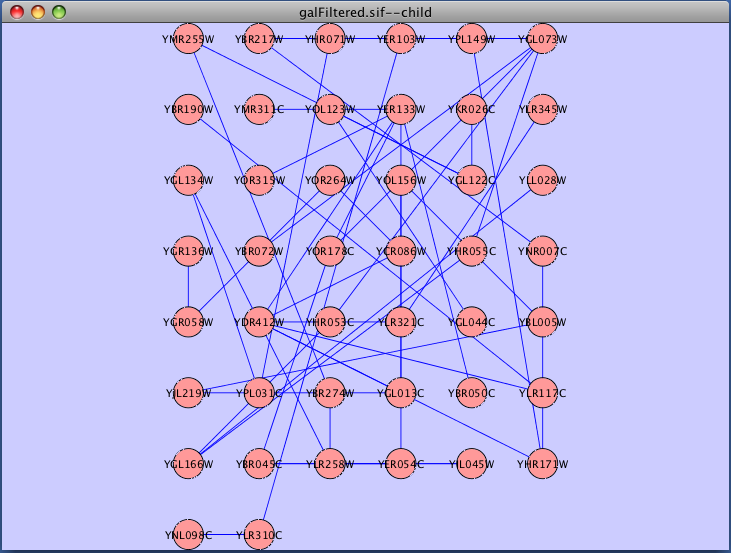
Force Directed Layout
The force-directed layout is a layout based on the "force-directed" paradigm. This layout is based on the algorithm implemented as part of the excellent prefuse toolkit provided by Jeff Heer. The algorithm is very fast and with the right parameters can provide a very pleasing layout. The Force Directed Layout will also accept a numeric edge attribute to use as a weight for the length of the spring, although this will often require more use of the Settings... dialog to achieve the best layout. This algorithm is available by selecting Layout → Cytoscape Layouts → Force-Directed Layout → (unweighted) or the edge attribute you want to use as a weight. A sample screen shot showing a portion of the galFiltered network provided in sample data is provided below:
Spring-Embedded Layout
The spring-embedded layout is based on a “force-directed” paradigm as implemented by Kamada and Kawai (1988). Network nodes are treated like physical objects that repel each other, such as electrons. The connections between nodes are treated like metal springs attached to the pair of nodes. These springs repel or attract their end points according to a force function. The layout algorithm sets the positions of the nodes in a way that minimizes the sum of forces in the network. This algorithm can be applied to the entire network or a portion of it by selecting the appropriate options from Layout → Cytoscape Layouts → Spring Embedded. A sample screen shot is provided below:
Attribute Circle Layout
The attribute circle layout is a quick, useful layout, particularly for small networks, that will locate all of the nodes in the network around a circle. The node order is determined by a user-selected node attribute. The result is that all nodes with the same value for that attribute are located together around the circle. Using Layout → Cytoscape Layouts → Attribute Circle Layout → attribute to put all nodes around a circle using attribute to position them. The sample screen shot below shows the a subset of the galFiltered network organized by node Degree.
Group Attributes Layout
The group attributes layout is similar to the attribute circle layout described above except that instead of a single circle with all of the nodes, each set of nodes that share the same value for the attribute are laid out in a separate circle. The same network shown above (subset of galFiltered) is shown below, using Layout → Cytoscape Layouts → Attribute Circle Layout → Degree.
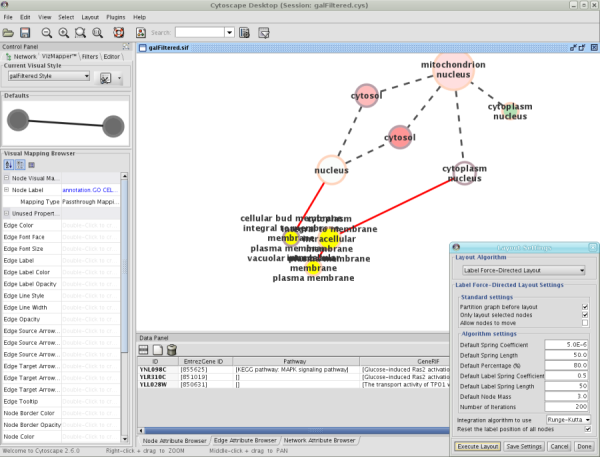
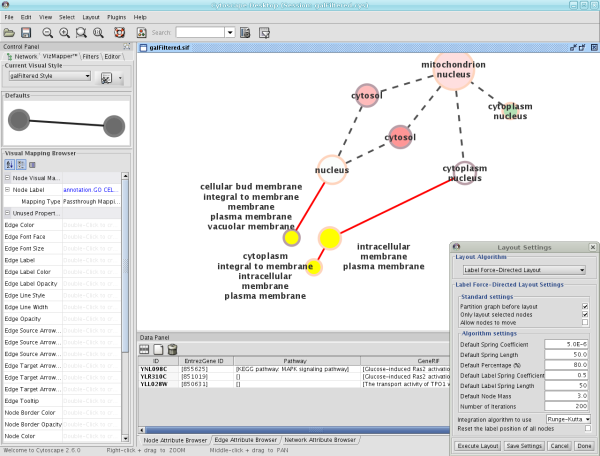
Label Force-Directed Layout
The Label Force-Directed Layout is an automatic label repositioning plugin that can be accessed through Layout → Cytoscape Layouts → Label Force-Directed Layout. The layout is designed to be applied to position labels after another network layout is run to position nodes. It is compatible with any node positioning layout. Specific numerical values that the algorithm makes use of can be set by the user through the Layout → Settings... menu. Through adjusting these values, you can control how much you want to spread the labels and/or nodes of the network by.
This label layout can be applied to the entire network, or only to those nodes that are selected (through enabling the Only layout selected nodes option). Upon the application of this plugin, node labels will be repositioned according to the Force-Directed algorithm, using the Default Label Spring Coefficient and Default Label Spring Length fields:
Default Label Spring Coefficient: The strength of the pull between the label of a node and the node itself. The distance between nodes and their labels will be closer to the Default Label Spring Length if this value is larger.
Default Label Spring Length: The desired length between the label and the node that the label belongs to.
Users can also choose to move the network nodes. This option can be activated by enabling the Allow nodes to move option. All network nodes (or just the selected ones) will be moved according to the Force-Directed layout algorithm, while making use of the Default Spring Coefficient, Default Spring Length, and Default Percentage (%) fields:
Default Spring Coefficient: The strength of the pull between a network node and its connected neighbours. The larger the value, the closer the distance between the two objects will be to the Default Spring Length value.
Default Spring Length: The desired length of an edge that is connecting two network nodes together.
Default Percentage (%): The percentage of the total Number of Iterations in which you would like the algorithm to move the network nodes as well, in addition to strictly moving the labels.
- For example: Number of Iterations = 200; Default Percentage (%) = 80. Then the network nodes will be moved for (80% / 100) * 200 = 160 of the 200 iterations.
Other algorithm settings include:
Default Node Mass: The mass that a default network node should take on. Nodes with a greater node mass require more strength to move.
Number of Iterations: The total number of times to run the algorithm for each execution of this label layout.
Reset the label position of all nodes: Reposition the label of each node to their default position.
Note: In order to yield the best layout for the labels, this plugin may need to be executed more than once.
Before
After
JGraph Layouts
Several other alignment algorithms, including a selection from the JGraph project (http://jgraph.sourceforge.net), are also available under the Layout menu.
Layout Parameters
Many layouts have adjustable parameters that are exposed through the Layouts → Settings... menu option. This will pop up the following dialog, which allows you to choose which layout algorithm settings to adjust. The settings presented vary by algorithm and only those algorithms that allow access to their parameters will appear in the drop-down menu at the top of the dialog. Once you've modified a parameter, clicking the "Execute Layout" button will apply the layout. This be repeated until a useful layout is achieved. Settings can be saved using the "Save Settings" button.
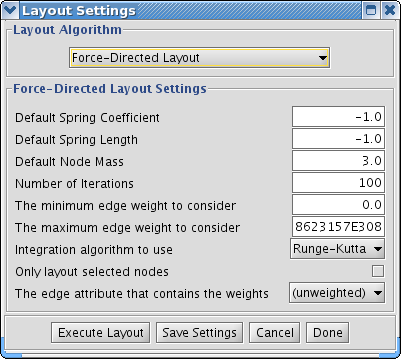
Manual Layout
The simplest method to manually organize a network is to click on a node and drag it. If you select multiple nodes, all of the selected nodes will be moved together.
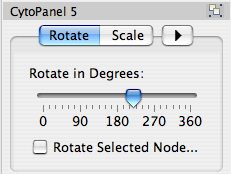
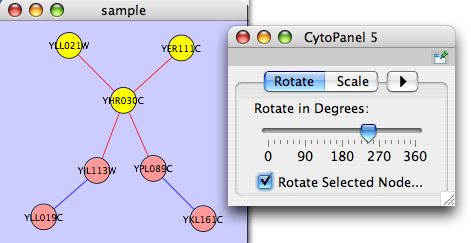
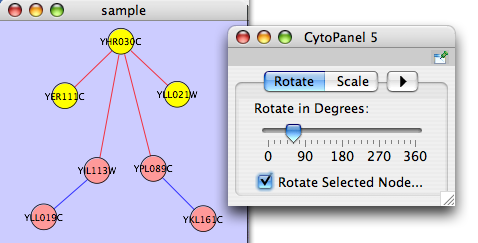
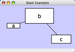
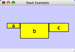
Rotate
Selecting the Layout → Rotate option will open the Rotate window in CytoPanel 5. This function will either rotate the entire network or a selected portion of the network. The image below shows a network with selected nodes rotated.
Before
After
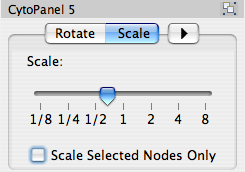
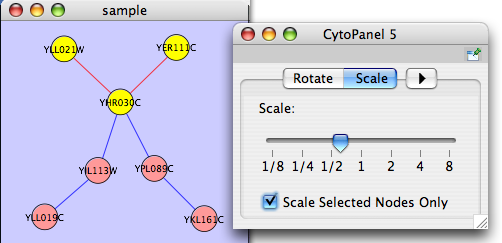
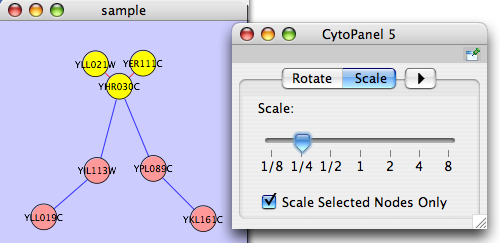
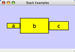
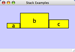
Scale
Selecting the Layout → Scale option will open the Scale window in CytoPanel 5. This function will scale the position of the entire network or of the selected portion of the network. Note that only the position of the nodes will be scaled, not the node sizes. Node size can be adjusted using the VizMapper. The image below shows selected nodes scaled.
Before
After
Align/Distribute/Stack
Selecting the Layout → Align/Distribute option will open the Align/Distribute/Stack window in CytoPanel 5. The Align buttons provide different options for either vertically or horizontally aligning selected nodes against a line. The differences are in what part of the node gets aligned, e.g. the center of the node, the top of the node, the left side of the node. The Distribute buttons evenly distribute selected nodes between the two most distant nodes along either the vertical or horizontal axis. The differences are again a function what part of the node is used as a reference point for the distribution. And the Stack buttons vertically or horizontally stack selected nodes with the full complement of alignment options. The table below provides a decription of what each button does.
Button |
Before |
After |
Description of Align Options |
|
|
|
Vertical Align Top - The tops of the selected nodes are aligned with the top-most node. |
|
|
|
Vertical Align Center - The centers of the selected nodes are aligned along a line defined by the midpoint between the top and bottom-most nodes. |
|
|
|
Vertical Align Bottom - The bottoms of the selected nodes are aligned with the bottom-most node. |
|
|
|
Horizontal Align Left - The left hand sides of the selected nodes are aligned with the left-most node. |
|
|
|
Horizontal Align Center - The centers of the selected nodes are aligned along a line defined by the midpoint between the left and right-most nodes. |
|
|
|
Horizontal Align Right - The right hand sides of the selected nodes are aligned with the right-most node. |
Button |
Before |
After |
Description of Distribute Options |
|
|
|
Vertical Distribute Top - The tops of the selected nodes are distributed evenly between the top-most and bottom-most nodes, which should stay stationary. |
|
|
|
Vertical Distribute Center - The centers of the selected nodes are distributed evenly between the top-most and bottom-most nodes, which should stay stationary. |
|
|
|
Vertical Distribute Bottom - The bottoms of the selected nodes are distributed evenly between the top-most and bottom-most nodes, which should stay stationary. |
|
|
|
Horizontal Distribute Left - The left hand sides of the selected nodes are distributed evenly between the left-most and right-most nodes, which should stay stationary. |
|
|
|
Horizontal Distribute Center - The centers of the selected nodes are distributed evenly between the left-most and right-most nodes, which should stay stationary. |
|
|
|
Horizontal Distribute Right - The right hand sides of the selected nodes are distributed evenly between the left-most and right-most nodes, which should stay stationary. |
Button |
Before |
After |
Description of Stack Options |
|
|
|
Vertical Stack Left - Vertically stacked below top-most node with the left-hand sides of the selected nodes aligned. |
|
|
|
Vertical Stack Center - Vertically stacked below top-most node with the centers of selected nodes aligned. |
|
|
|
Vertical Stack Right - Vertically stacked below top-most node with the right-hand sides of the selected nodes aligned. |
|
|
|
Horizontal Stack Top - Horizontally stacked to the right of the left-most node with the tops of the selected nodes aligned. |
|
|
|
Horizontal Stack Center - Horizontally stacked to the right of the left-most node with the centers of selected nodes aligned. |
|
|
|
Horizontal Stack Bottom - Horizontal Stack Center - Horizontally stacked to the right of the left-most node with the bottoms of the selected nodes aligned. |
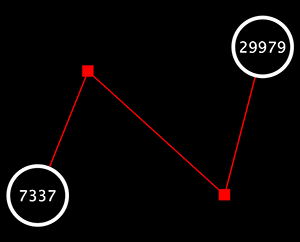
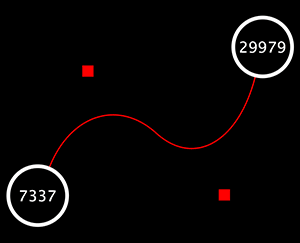
Edge Handles
This is a little-known feature! If you select an edge and then Ctrl-left-click on the edge, an edge "handle" will appear. This handle can be used to change the shape of the line. To remove a handle, simply Ctrl-left-click on the handle again.
Straight Edge |
Smooth Edge |
|
|
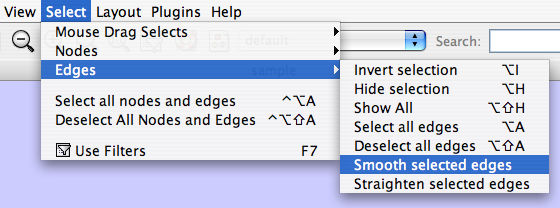
Two other options under Select → Edges provide further control: "Smooth Selected Edges" turns an edge consisting of line segments into a smoothed bezier curve, and "Straighten Selected Edges" turns a curved edge back into line segments.
Node Movement and Placement
In addition to the ability to click on a node and drag it to a new position, Cytoscape now has the ability to move nodes using the arrow keys on the keyboard. By selecting one or more nodes using the mouse and clicking one of the arrow keys (←, →, ↑, ↓) the selected nodes will move one pixel in the chosen direction. If an arrow key is pressed while holding the Shift key down, the selected nodes will 10 pixels in the chosen direction.
Mouse movement has also been enhanced. If the Shift key is held down while dragging a node, the node will only move horizontally, vertically, or along a 45 degree diagonal.