|
Size: 2642
Comment:
|
Size: 3217
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 3: | Line 3: |
| In this chapter, you will learn how to use Cytoscape from command line and scripts. | In this chapter, you will learn how to use Cytoscape from command line and scripts. These features replace ''Scripting'' module in the past versions of Cytoscape. |
| Line 5: | Line 5: |
| ==== Topics ==== | === Topics === |
| Line 7: | Line 7: |
|
* Commands * RESTful API * Command REST API * cyREST |
* '''''Commands''''' * '''RESTful API''' * '''Command REST API''' * '''cyREST''' |
| Line 18: | Line 18: |
| === Options for Programmatic Access === | == Options for Programmatic Access == |
| Line 21: | Line 21: |
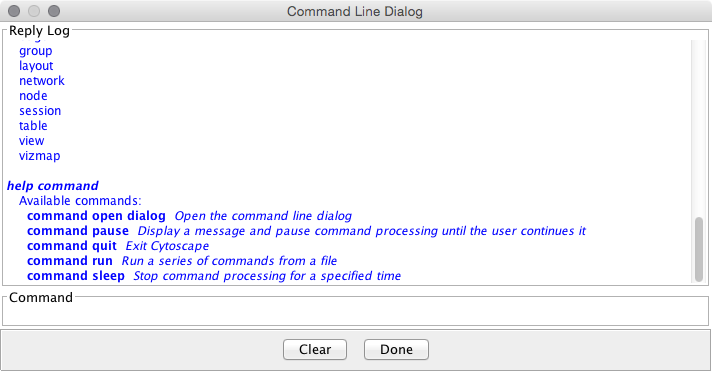
| ==== Commands ==== | === Commands === |
| Line 26: | Line 26: |
| '''''Commands''''' is the built-in Cytoscape feature to automate your workflow as a simple script. You can learn more about this feature in this section: | '''''Commands''''' is the built-in Cytoscape feature to automate your workflow as simple script. You can learn more about this feature in this section: |
| Line 31: | Line 31: |
|
===== Sample Scripts ===== {{{ (TBD) }}} ==== RESTful API ==== |
=== RESTful API === |
| Line 40: | Line 34: |
|
===== Commands REST API ===== In addition to running scripts, Commands have REST API to access the |
==== REST API for Commands ==== In addition to running Command scripts, Command module has REST API to access those |
| Line 47: | Line 41: |
| * Terminal or [[|iTerm]] (For Mac) | * Terminal or [[https://www.iterm2.com/|iTerm2]] (For Mac) |
| Line 72: | Line 66: |
| ===== cyREST ===== | ==== cyREST ==== |
| Line 76: | Line 70: |
| '''[[http://apps.cytoscape.org/apps/cyrest|cyREST]] is a language-agnostic, programmer-friendly RESTful API module for Cytoscape'''. If you want to build your own workflow with [[http://www.r-project.org/|R]], [[https://www.python.org/|Python]] or other programming languages along with Cytoscape, this is the option for you. | |
| Line 77: | Line 72: |
| cyREST is available from |
{{attachment:jupyter.png}} Currently, cyREST is available as an App for Cytoscape 3.2.1 and later. * [[http://apps.cytoscape.org/apps/cyrest|cyREST App Store page]]. |
Programmatic Access to Cytoscape Features
In this chapter, you will learn how to use Cytoscape from command line and scripts. These features replace Scripting module in the past versions of Cytoscape.
Topics
Commands
RESTful API
Command REST API
cyREST
Background
Cytoscape has intuitive graphical user interface for interactive network data integration, analysis, and visualization. It is a great way for exploratory data analysis, but what happens if you have hundreds of data files or need to ask someone to execute your data analysis workflows? It is virtually impossible to apply same operations to hundreds of networks manually with GUI, and more importantly, although you can save your results as session files, you cannot save your workflows if your analysis is based on point-and-click GUI operation. Recording workflows is the key feature for reproducible data analysis, but it was a missing feature in past versions for Cytoscape. The latest version of Cytoscape has several options to script and automate your workflows.
Options for Programmatic Access
There are two ways to access Cytoscape programmatically: Commands and RESTful API.
Commands
Commands is the built-in Cytoscape feature to automate your workflow as simple script. You can learn more about this feature in this section:
RESTful API
In some cases, you may need to use fully featured programming languages, such as Python, R, Ruby, or JavaScript,
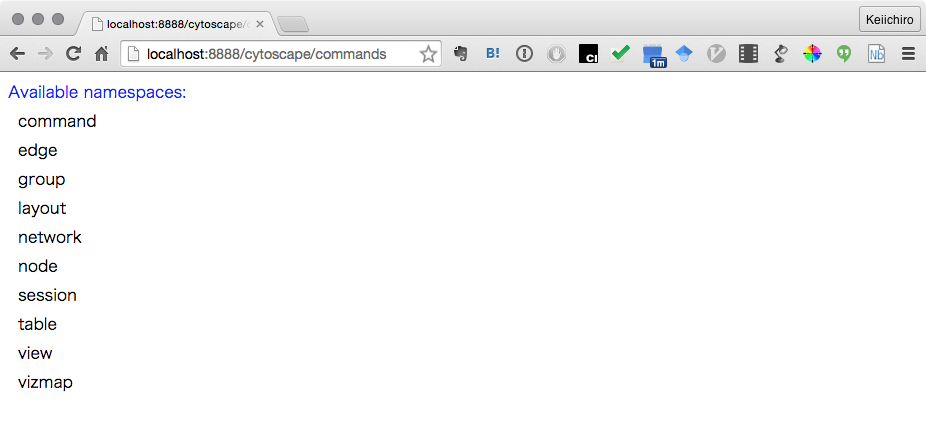
REST API for Commands
In addition to running Command scripts, Command module has REST API to access those
By default, this feature is disables. To run REST server for Commands, please follow these steps:
- Open a terminal session:
PowerShell (For windows)
Terminal or iTerm2 (For Mac)
- Terminal (For Linux)
- Start Cytoscape from command-line. You need to specify port number as a parameter (In this example, port 8888 will be opened for Command):
- For Mac/Linux
./cytoscape.sh -R 8888
For Windows./cytoscape.bat -R 8888
- For Mac/Linux
- Open the following URL with your web browser:
http://localhost:8888/cytoscape/commands
- If yo see list of available commands, you are ready to use Command API
cyREST

cyREST is a language-agnostic, programmer-friendly RESTful API module for Cytoscape. If you want to build your own workflow with R, Python or other programming languages along with Cytoscape, this is the option for you.
Currently, cyREST is available as an App for Cytoscape 3.2.1 and later.
Debugging your workflows
(TBD)