|
← Revision 1 as of 2013-12-16 23:01:02
Size: 1176
Comment:
|
← Revision 2 as of 2013-12-16 23:11:39 →
Size: 1177
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 12: | Line 12: |
| To run !NetworkAnalyzer, select '''Tools → NetworkAnalyzer → Network Analysis → Analyze Network'''. | To run !NetworkAnalyzer, select '''Tools → !NetworkAnalyzer → Network Analysis → Analyze Network'''. |
NetworkAnlayzer computes a comprehensive set of topological parameters for undirected and directed networks, including:
- Number of nodes, edges and connected components.
- Network diameter, radius and clustering coefficient, as well as the characteristic path length.
- Charts for topological coefficients, betweenness, and closeness.
- Distributions of degrees, neighborhood connectiveness, average clustering coefficients, shortest path lengths, number of shared neighbors and stress centrality.
NetworkAnalyzer also constructs the intersection, union and difference of two networks. It supports the extraction of connected components as separate networks and the removal of self-loops.
Analyze Network
To run NetworkAnalyzer, select Tools → NetworkAnalyzer → Network Analysis → Analyze Network.
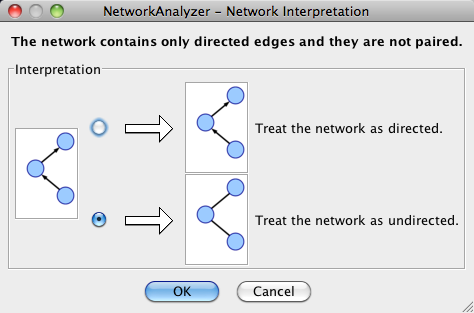
The NetworkAnalyzer will determine whether your network contains directed or undirected edges. At this point, you can choose to ignore edge direction information.
Explanation of analysis parameters: http://med.bioinf.mpi-inf.mpg.de/netanalyzer/help/2.7/index.html#simple