|
← Revision 10 as of 2013-12-11 00:00:03
Size: 4181
Comment:
|
← Revision 11 as of 2013-12-11 22:01:57 →
Size: 4182
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 31: | Line 31: |
| == Adding or Removing Links == | == Adding and Removing Links == |
Linkout provides a mechanism to link nodes and edges to external web resources within Cytoscape. Right-clicking on a node or edge in Cytoscape opens a popup menu with a list of web links.
The external links are specified in a linkout.props file which is internal to Cytoscape. The defaults include a number of links such as Entrez, SGD, iHOP, and Google, as well as a number of species-specific links. In addition to the default links, users can customize the External Links menu and add (or remove) links by editing the linkout properties (found under Edit → Preferences → Properties...).
External links are listed as ‘key’-‘value’ pairs in the linkout.props file where key specifies the name of the link and value is the search URL. The LinkOut menus are organized in a hierarchical structure that is specified in the key. Linkout key terms specific for nodes start with the keyword nodelinkouturl, for edges this is edgelinkouturl.
For example, the following entry:
nodelinkouturl.Model Organism DB.SGD (yeast)=http://www.yeastgenome.org/cgi-bin/locus.fpl?locus=%ID%
places the SGD link under the Model Organism DB submenu. This link will appear in Cytoscape as:
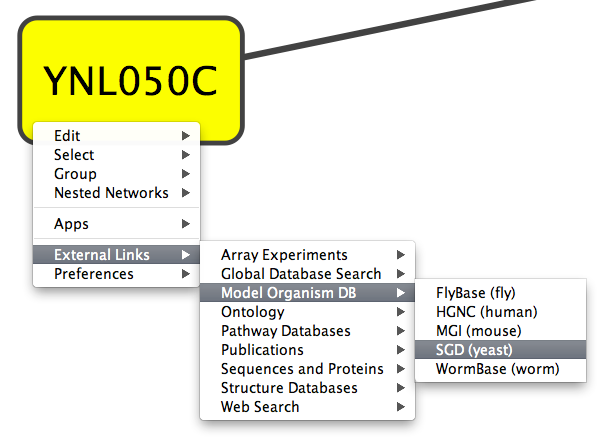
In a similar fashion one can add new submenus.
The %ID% string in the URL is a place-holder for the node label. When the popup menu is generated this marker is substituted with the node label. In the above example, the generated SGD link for the YNL050C protein is:
http://www.yeastgenome.org/cgi-bin/locus.fpl?locus=YNL050C
If you want to query based on a different column, you need to specify a different node label using Styles.
For edges the mechanism is much the same; however here the placeholders %ID1% and %ID2% reflect the source and target node label respectively.
Currently there is no mechanism to check whether the constructed URL query is correct and if the node label is meaningful. Similarly, there is no ID mapping between various identifiers. For example, a link to NCBI Entrez from a network that uses Ensembl gene identifiers as node labels will produce a link to Entrez using the Ensembl ID, which results in an incorrect link. It is the user's responsibility to ensure that the node label that is used as the search term in the URL link will result in a meaningful link.
Adding and Removing Links
The default links are defined in a linkout.props file contained inside the Linkout JAR bundle under the framework/system/org/cytoscape/linkout-impl subdirectory of the Cytoscape installation. These links are normal Java properties and can be edited by going to Edit → Preferences → Properties... and selecting linkout from the box (shown below). Linkouts can be modified, added or removed using this dialog; however, note that the modifications would not be stored in the file. To change a URL permanently, you would need to edit the linkout.props file directly.
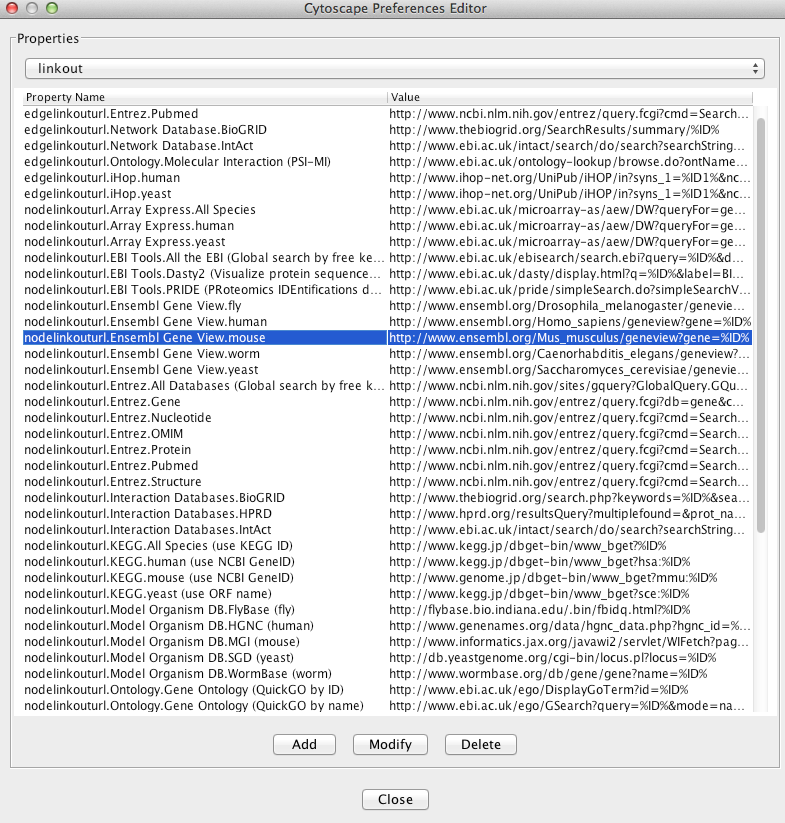
In addition, new links can be defined when starting Cytoscape from command line by specifying individual properties. The formatting of the command is cytoscape.sh -P [context_menu_definition]=[link] . context_menu_definition specifies the context menu for showing the linkout menu item. The structure of this definition is "." separated and the first item needs to be either nodelinkouturl or edgelinkouturl. The former will add the linkout item as a node context menu and the latter will add it as an edge context menu. The rest of the definition would define the hierarchy of the menu.
For instance this command:
cytoscape.sh -P nodelinkouturl.yeast.SGD=http://db.yeastgenome.org/cgi-bin/locus.pl?locus\=%ID%
will add this menu item:
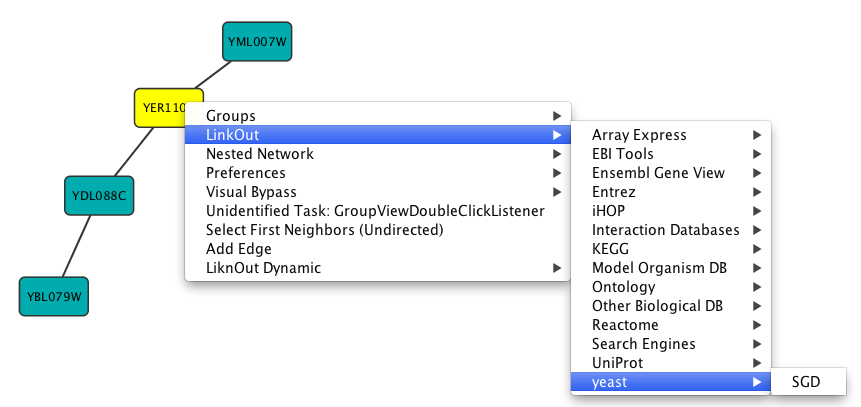
To remove a link from the menu, simply delete the property using Edit → Preferences → Properties... and selecting commandline. Linkouts added in the command line will be available for the running instance of Cytoscape.