|
← Revision 29 as of 2014-02-06 22:49:21
Size: 6663
Comment:
|
← Revision 30 as of 2014-02-08 01:21:50 →
Size: 7192
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 21: | Line 21: |
|
If not already installed on your computer, download and install Java SE 6. Cytoscape functionality has not been verified yet with SE 7. Link to download SE 6: [[http://java.sun.com/javase/downloads/index.jsp|Java SE 6]] |
64-bit Java Runtime Environment (JRE) is recommended – the latest Oracle distribution can be found at [[http://java.sun.com/javase/downloads/index.jsp|Latest JRE]]. Cytoscape has been tested with Java 6 and 7, but not later versions. For 64-bit Windows systems, the Windows x64 version is recommended. For Linux systems, Oracle Java 7 has caused Cytoscape crashes on some platforms, and OpenJDK7 (http://openjdk.java.net) is a good alternative. For Mac systems, the Cytoscape installer automatically triggers Java6 to install if no other Java version is present. |
| Line 25: | Line 23: |
| '''Warning:''' The default Java download for Windows is 32 bits. If you plan to run 64 bit Cytoscape, you must download 64 bit Java instead. |
Note that if you are a Windows user and you choose to download Java from http://java.net, beware that the default download from this site is 32-bit JRE. If your Windows is 64-bits, the 64-bit JRE is recommended. For additional information, select the Release Notes button on the Cytoscape web site (http://cytoscape.org). |
Cytoscape is a Java application verified to run on the Linux, Windows, and Mac OS X platforms. Although not officially supported, other UNIX platforms such as Solaris or FreeBSD may run Cytoscape if Java version 6 or later is available for the platform.
System requirements
The system requirements for Cytoscape depend on the size of the networks the user wants to load, view and manipulate.
|
Small Network Visualization |
Large Network Analysis/Visualization |
Processor |
1GHz |
As fast as possible |
Memory |
512MB |
2GB+ |
Graphics Card |
Integrated video |
High-end graphics Card |
Monitor |
XGA (1024X768) |
Wide or Dual Monitor |
Specific system requirements, limitations, and configuration options apply to each platform, as described in the Release Notes available on the http://cytoscape.org website.
Directory Location
For the application to work properly, all files should be left in the directory in which they were unpacked. The core Cytoscape application assumes this directory structure when looking for the various libraries needed to run the application. For more information on program directories, see the directory table below.
Getting Started
Install Java
64-bit Java Runtime Environment (JRE) is recommended – the latest Oracle distribution can be found at Latest JRE. Cytoscape has been tested with Java 6 and 7, but not later versions. For 64-bit Windows systems, the Windows x64 version is recommended. For Linux systems, Oracle Java 7 has caused Cytoscape crashes on some platforms, and OpenJDK7 (http://openjdk.java.net) is a good alternative. For Mac systems, the Cytoscape installer automatically triggers Java6 to install if no other Java version is present.
Note that if you are a Windows user and you choose to download Java from http://java.net, beware that the default download from this site is 32-bit JRE. If your Windows is 64-bits, the 64-bit JRE is recommended.
For additional information, select the Release Notes button on the Cytoscape web site (http://cytoscape.org).
Install Cytoscape
Downloading and installing
There are a number of options for downloading and installing Cytoscape. See the download page at the http://cytoscape.org website for all options.
- Automatic installation packages exist for Windows, Mac OS X, and Linux platforms.
- You can install Cytoscape from a compressed archive distribution.
- You can build Cytoscape from the source code.
You can check out the latest and greatest software from our Git repository.
Cytoscape installations (regardless of platform) containing the following files and directories:
File/Directory |
Description |
p/Cytoscape_v3.1.0 |
Cytoscape program files, startup scripts, and default location for session files |
p/Cytoscape_v3.1.0/Cytoscape.vmoptions |
Cytoscape memory configuration settings |
p/Cytoscape_v3.1.0/sampleData |
Preset networks as described in the embedded README.txt file |
p/Cytoscape_v3.1.0/framework |
Cytoscape program files |
u/CytoscapeConfiguration |
Cytoscape properties and program cache files |
u/CytoscapeConfiguration/cytoscape3.props |
Cytoscape configuration settings |
The p/ directory signifies the program directory, which varies from platform to platform.
The u/ directory signifies the user's home directory, which varies from user to user and from platform to platform.
A quick note on upgrading
If you have a previous installation you have two options:
Starting with a clean slate. For this you should delete your previous installation directory and the CytoscapeConfiguration directory (see below for the location of this directory).
Just keep what you have and simply pick a distinct, new directory for installation. In the unlikely event that you should encounter any problem, delete the .props files in your CytoscapeConfiguration directory. If that doesn't help try deleting the CytoscapeConfiguration directory. This latter step will cause you to lose all of the apps that you have installed via the App Store, so only do that if you are having problems or if you don't mind reinstalling your apps. The core apps will not be affected by this step.
Launch the application
As with any application: double-click on the icon created by the installer or by running cytoscape.sh from the command line (Linux or Mac OS X) or double-clicking cytoscape.bat (Windows).
After launching Cytoscape a window will appear that looks like this:
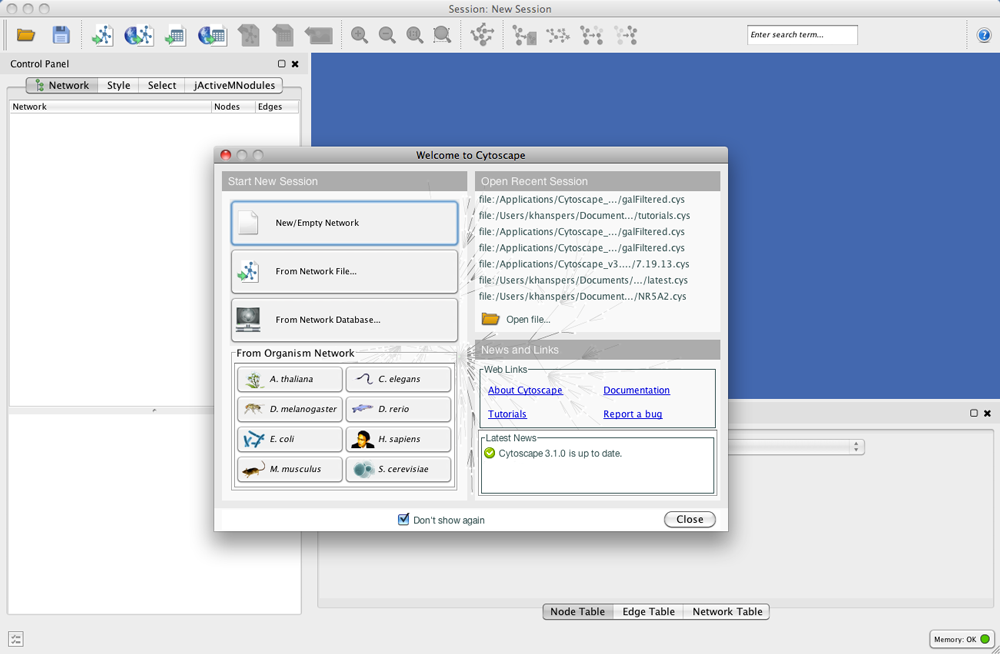
If your Cytoscape window does not resemble this, further configuration may be required. Consult the Release Notes available on the http://cytoscape.org website.
Note on Memory Consumption
For most regular users Cytoscape will estimate the proper amount of memory, if so; you can skip this section and continue to the Quick Tour.
Cytoscape will display the current memory usage during runtime in the lower right corner of the main interface. You can click on the Memory button at any time to access an option to Free Unused Memory. While most users won't need to use this option, it can be useful for users who have multiple large networks loaded.
Overall Memory Size for Cytoscape
By default, Cytoscape uses an estimate for memory allocation based on your operating system, system architecture (32 or 64 bit), and installed memory. To increase the maximum memory size for Cytoscape, you can specify it in a file, residing in the same directory as the Cytoscape executable, called Cytoscape.vmoptions with one option per line and lines separated by linefeeds. The last line must also be followed by a linefeed. The one exception to this rule is the MacOS platform if you are launching Cytoscape by clicking on the Cytoscape icon. (In that case you will have to edit the .../Cytoscape.app/Contents/Info.plist file instead!) For example, if you want to assign 4GB of memory, create the Cytoscape.vmoptions file containing the single line (Do not forget the linefeed at the end of the line!):
-Xmx4GB
Stack Size
There is one more option related to memory allocation. Some of the functions in Cytoscape use larger stack space (a temporary memory for some operations, such as Layout). Since this value is set independently from the Xmx value above, sometimes Layout algorithms fails due to the out of memory error. To avoid this, you can set larger heap size for Cytoscape tasks by using the taskStackSize option in the cytoscape3.props file (located in the CytoscapeConfiguration directory). This can be edited within Cytoscape using the Preferences Editor (Edit-Preferences-Properties...) - look for taskStackSize. The value should be specified in bytes.