|
← Revision 23 as of 2014-01-23 20:42:49
Size: 6493
Comment:
|
← Revision 24 as of 2016-04-26 22:25:34
Size: 61
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 1: | Line 1: |
|
== What is Cytoscape.js? == {{attachment:jsWebSite.png}} [[http://cytoscape.github.io/cytoscape.js/|Cytoscape.js]] is a '''!JavaScript library''' for interactive network visualization. It is a building block for web applications and is '''NOT''' a complete web application. If you have network data sets and want to share visualizations created with Cytoscape, you can build your own website using Cytoscape.js and this new '''Export to Cytoscape.js''' feature. === Examples === * A network visualized with Cytoscape 3.1.0 {{attachment:sampleOriginal.png}} * Same network exported to Cytoscape.js (Rendered with Google Chrome and Cytoscape.js 2.0.3) {{attachment:sampleExport.png}} * [[http://chianti.ucsd.edu/~kono/dist/|Interactive example (galFiltered.sif rendered with Cytoscape.js 2.0.3)]] === Differences and Shared Concepts === Although Cytoscape and Cytoscape.js are two completely independent software packages, they are sharing higher level concepts. The following is the list of similarities and differences between the two: ==== Cytoscape ==== * '''Desktop application''' for network visualization written in [[http://www.java.com/|Java programming language]] * Needs desktop or laptop computers to run * Users have to install Java runtime * High performance application for large scale network analysis and visualization * Expandable by [[http://apps.cytoscape.org/|Apps]] * Use '''Styles''' to map data to network properties, such as node color, edge width, node shape, etc. ==== Cytoscape.js ==== * A '''[[http://en.wikipedia.org/wiki/JavaScript|JavaScript library]]''' for network visualization, '''NOT''' a complete web application nor mobile app * Runs on most of modern web browsers, including tablets and smart phones * No plugins are required to run. Modern web browser is the only requirement * Need to write code to set up your web site or web application * Expandable by [[http://cytoscape.github.io/cytoscape.js/#extensions|Extensions]] * Use '''CSS-based Styles''' to map data to network properties In a long term, Cytoscape and Cytoscape.js will be more integrated, and as the first step Cytoscape now supports reading and writing Cytoscape.js network/table JSON files. In addition, Cytoscape can convert ''Styles'' to Cytoscape.js Style object. == Data Exchange between Cytoscape and Cytoscape.js == Since Cytoscape.js is a !JavaScript library, its basic data exchange format is [[http://www.json.org/|JSON (JavaScript Object Notation)]]. All of these import/export functions are part of standard Cytoscape user interface, and you can read/write Cytoscape.js JSON files just like other standard files such as SIF. === Export Network and Table to Cytoscape.js === Cytoscape.js stores network data (graph) and its data table in the same object. Cytoscape writes such data complex as JSON, i.e., both network and data tables will be exported as a single JSON file. You can select a network and export it from '''File → Export → Network'''. Cytoscape only supports one of the Cytoscape.js supported JSON formats, which is: {{{ { elements:{ nodes:[], edges:[] } } }}} '''SUID''' will be used as unique identifier for nodes and edges in the JSON. For more information about this format, please visit [[http://cytoscape.github.io/cytoscape.js/|Cytoscape.js web site]]. ==== Important Note about Data Compatibility ==== Cytoscape creates JSON file directly from data table and tries to extract as much data as possible from the original table. However, since table column names will be directly converted into JavaScript variable names, invalid characters will be replaced by underscore (_): * Original Data Table Column Names: {{{ Gene Name KEGG.pathway }}} * The Names above will be replaced to: {{{ Gene_Name KEGG_pathway }}} You should be careful when you plan to use this feature for data roundtrip: from Cytoscape to Cytoscape.js back to Cytoscape. For such use cases, '''we strongly recommend to use JavaScript-safe characters only in your table column names'''. Naming your columns only with alphanumeric characters and underscore (_) is the best practice. (For actual data entries, all characters are allowed. This restriction is only for table column names.) === Export Styles to Cytoscape.js === Cytoscape and Cytoscape.js are sharing a concept called '''Style'''. This is a collection of mappings from data point to network property. Cytoscape can export its Styles into CSS-based Cytoscape.js JSON. You can export ''all Styles into one JSON file'' from '''File → Export → Style''' and select Cytoscape.js JSON as its format. ==== Limitations ==== Cytoscape.js does not support all of Cytoscape Network Properties. Those properties will be ignored or simplified when you export to JSON Style file. Currently, the following Visual Properties will not be exported to Cytoscape.js JSON: * Custom Graphics and its positions * Edge Bends * Tooltips * Node Label Width * Node Border Line Type * Arrow Colors (they are always same as edge color) ==== Cytoscape.js and Cytoscape compatibility charts ==== The following two charts detail the compatibility of specific Cytoscape features in Cytoscape.js. [[attachment:CS-web_vs_CS-1_small.png|{{attachment:CS-web_vs_CS-1.png||width=100}}]] [[attachment:CS-web_vs_CS-2_small.png|{{attachment:CS-web_vs_CS-2.png||width=100}}]] === Import Cytoscape.js data into Cytoscape === Cytoscape.js network JSON files can be loaded from standard Cytoscape file menu: '''File → Import → Network →'''. Both File and URL are supported. == Build Your Own Web Application with Cytoscape.js == Although Cytoscape can export networks, tables, and Style as Cytoscape.js-compatible JSON, users have to write some JavaScript code to visualize the data files with Cytoscape.js. Details of web application development with Cytoscape.js is beyond the scope of this document. If you need examples and tutorials about web application development with Cytoscape.js, please visit the following web site: * https://github.com/cytoscape/cyjs-sample/wiki === Questions? === If you have questions and comments about web application development with Cytoscape and Cytoscape.js, please send yours to [[https://groups.google.com/forum/#!forum/cytoscape-discuss|our mailing list]]. |
#DEPRECATED <<Include(Cytoscape_3/UserManual/Deprecated)>> |
This is a legacy document
This page has been deprecated and is no longer updated. The current version of the Cytoscape manual can be found at http://manual.cytoscape.org/
What is Cytoscape.js?
Cytoscape.js is a JavaScript library for interactive network visualization. It is a building block for web applications and is NOT a complete web application. If you have network data sets and want to share visualizations created with Cytoscape, you can build your own website using Cytoscape.js and this new Export to Cytoscape.js feature.
Examples
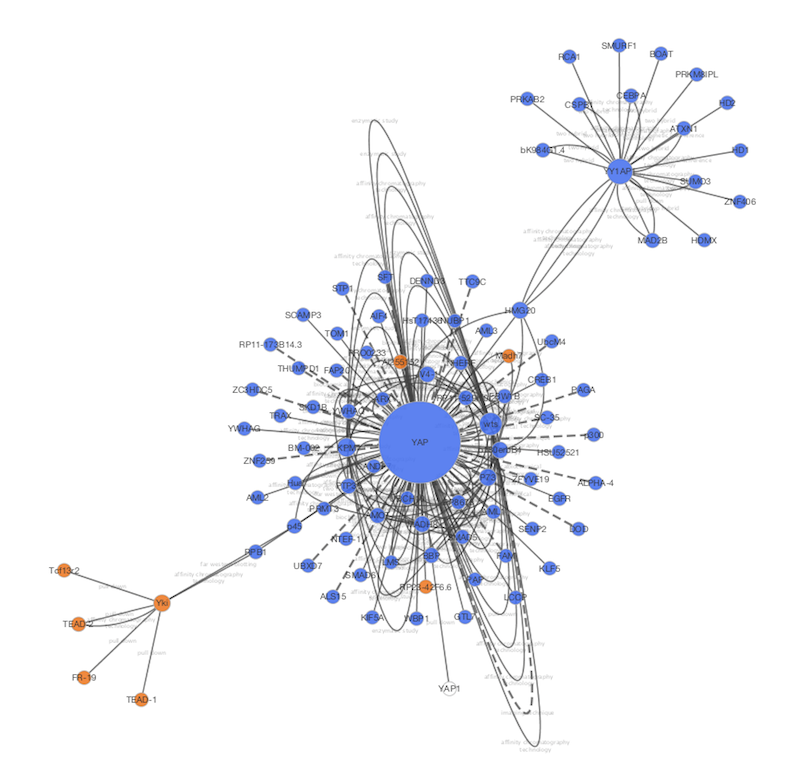
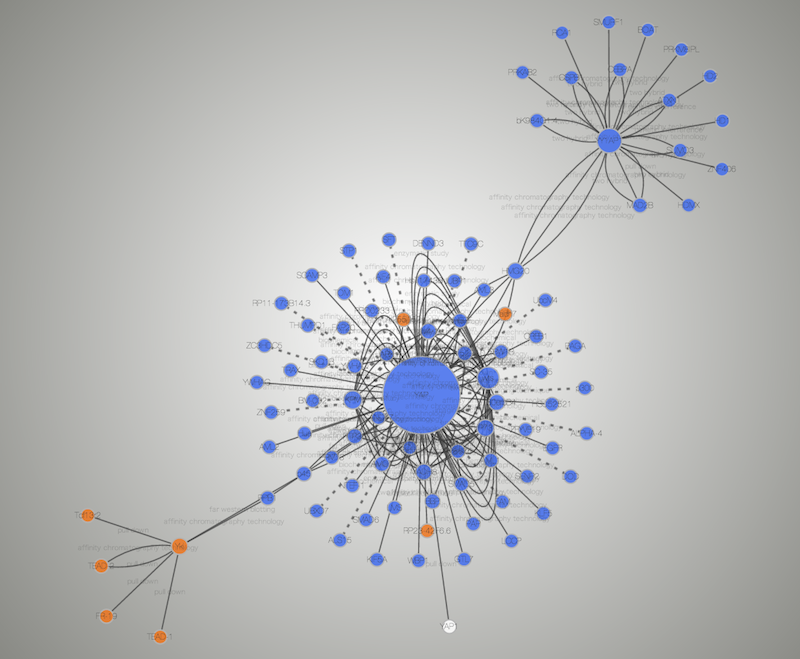
- A network visualized with Cytoscape 3.1.0
- Same network exported to Cytoscape.js (Rendered with Google Chrome and Cytoscape.js 2.0.3)
Interactive example (galFiltered.sif rendered with Cytoscape.js 2.0.3)
Differences and Shared Concepts
Although Cytoscape and Cytoscape.js are two completely independent software packages, they are sharing higher level concepts. The following is the list of similarities and differences between the two:
Cytoscape
Desktop application for network visualization written in Java programming language
- Needs desktop or laptop computers to run
- Users have to install Java runtime
- High performance application for large scale network analysis and visualization
Expandable by Apps
Use Styles to map data to network properties, such as node color, edge width, node shape, etc.
Cytoscape.js
A JavaScript library for network visualization, NOT a complete web application nor mobile app
- Runs on most of modern web browsers, including tablets and smart phones
- No plugins are required to run. Modern web browser is the only requirement
- Need to write code to set up your web site or web application
Expandable by Extensions
Use CSS-based Styles to map data to network properties
In a long term, Cytoscape and Cytoscape.js will be more integrated, and as the first step Cytoscape now supports reading and writing Cytoscape.js network/table JSON files. In addition, Cytoscape can convert Styles to Cytoscape.js Style object.
Data Exchange between Cytoscape and Cytoscape.js
Since Cytoscape.js is a JavaScript library, its basic data exchange format is JSON (JavaScript Object Notation). All of these import/export functions are part of standard Cytoscape user interface, and you can read/write Cytoscape.js JSON files just like other standard files such as SIF.
Export Network and Table to Cytoscape.js
Cytoscape.js stores network data (graph) and its data table in the same object. Cytoscape writes such data complex as JSON, i.e., both network and data tables will be exported as a single JSON file. You can select a network and export it from File → Export → Network.
Cytoscape only supports one of the Cytoscape.js supported JSON formats, which is:
{
elements:{
nodes:[],
edges:[]
}
}SUID will be used as unique identifier for nodes and edges in the JSON. For more information about this format, please visit Cytoscape.js web site.
Important Note about Data Compatibility
Cytoscape creates JSON file directly from data table and tries to extract as much data as possible from the original table. However, since table column names will be directly converted into JavaScript variable names, invalid characters will be replaced by underscore (_):
- Original Data Table Column Names:
Gene Name KEGG.pathway
- The Names above will be replaced to:
Gene_Name KEGG_pathway
You should be careful when you plan to use this feature for data roundtrip: from Cytoscape to Cytoscape.js back to Cytoscape. For such use cases, we strongly recommend to use JavaScript-safe characters only in your table column names. Naming your columns only with alphanumeric characters and underscore (_) is the best practice. (For actual data entries, all characters are allowed. This restriction is only for table column names.)
Export Styles to Cytoscape.js
Cytoscape and Cytoscape.js are sharing a concept called Style. This is a collection of mappings from data point to network property. Cytoscape can export its Styles into CSS-based Cytoscape.js JSON.
You can export all Styles into one JSON file from File → Export → Style and select Cytoscape.js JSON as its format.
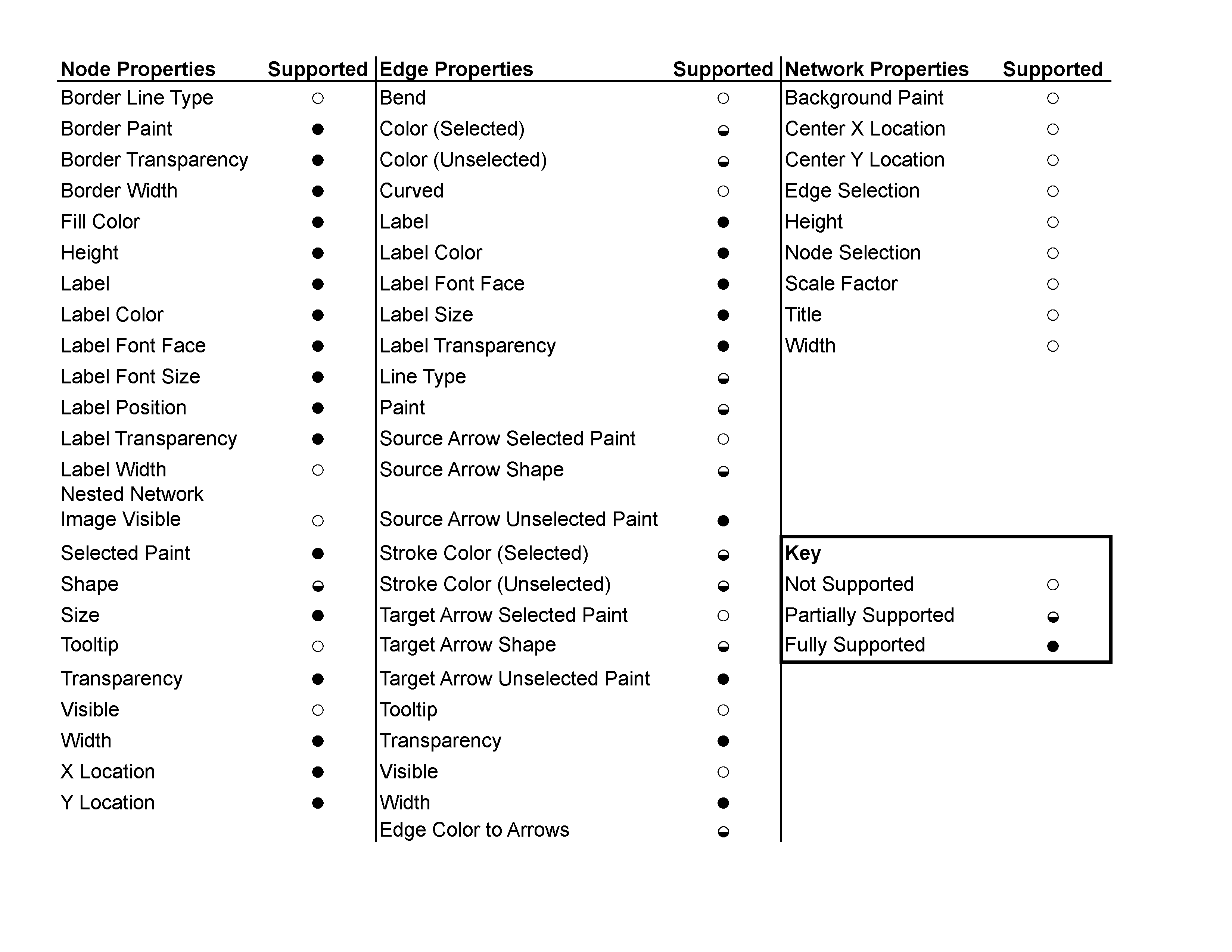
Limitations
Cytoscape.js does not support all of Cytoscape Network Properties. Those properties will be ignored or simplified when you export to JSON Style file.
Currently, the following Visual Properties will not be exported to Cytoscape.js JSON:
- Custom Graphics and its positions
- Edge Bends
- Tooltips
- Node Label Width
- Node Border Line Type
- Arrow Colors (they are always same as edge color)
Cytoscape.js and Cytoscape compatibility charts
The following two charts detail the compatibility of specific Cytoscape features in Cytoscape.js.
Import Cytoscape.js data into Cytoscape
Cytoscape.js network JSON files can be loaded from standard Cytoscape file menu: File → Import → Network →. Both File and URL are supported.
Build Your Own Web Application with Cytoscape.js
Although Cytoscape can export networks, tables, and Style as Cytoscape.js-compatible JSON, users have to write some JavaScript code to visualize the data files with Cytoscape.js. Details of web application development with Cytoscape.js is beyond the scope of this document. If you need examples and tutorials about web application development with Cytoscape.js, please visit the following web site:
Questions?
If you have questions and comments about web application development with Cytoscape and Cytoscape.js, please send yours to our mailing list.