|
← Revision 4 as of 2013-12-04 23:46:40
Size: 1477
Comment:
|
← Revision 5 as of 2013-12-05 01:05:14 →
Size: 2641
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 1: | Line 1: |
| The '''Command Line Tool''' provides a simple command-line interface to Cytoscape using the CyCommands API. Any app that registers commands will be available through the Command Tool. The Command Tool provides two main functions: first, a Command Line Dialog is available from '''Tools → Command Line Dialog''', that allows the user to type commands into Cytoscape and see the results in a "Reply Log". | The '''Command Line Tool''' provides a simple command-line interface to Cytoscape using the Commands API. Any app that registers commands will be available through the Command Tool. The Command Tool provides two main functions: first, a Command Line Dialog is available from '''Tools → Command Line Dialog''', that allows the user to type commands into Cytoscape and see the results in a "Reply Log". |
| Line 9: | Line 9: |
| The Command Tool uses the CyCommand API to provide help. "help" by itself will list all of the command classes (or namespaces) and "help " followed by a namespace will list all of the commands supported by that namespace. Details of a specific command are available by typing "help " followed by the namespace and command (e.g. "help layout force-directed"). The Command Tool registers the "commandtool" namespace and supports a single command: run, which takes a file argument. Here is the help for the commandtool run command from commandtool: | Cytoscape commands consist of three parts: a command class, or namespace; a command within that namespace; and a series of arguments or options provided as a series of '''name=value''' pairs. For example, to import an XGMML format file from the '''Command Line Dialog''' or a command script, you would use: |
| Line 11: | Line 11: |
|
help commandtool run commandtool run: Run a command script from a file Arguments: [file=value] |
{{{ network import file filePath="path-to-file" }}} where ''network'' is the namespace, ''import file'' is the command, and there is only one argument: ''filePath="path-to-file"''. If there were more arguments they would appear on the same line separated by spaces. The Command Tool also uses the Command API to provide help. "help" by itself will list all of the command classes (or namespaces) and "help " followed by a namespace will list all of the commands supported by that namespace. Details of a specific command are available by typing "help " followed by the namespace and command (e.g. "help layout force-directed"). The Command Tool registers the "command" namespace and supports a single command: run, which takes a file argument. Here is the help for the command run command from the command namespace: {{{ help command run command run file=<File> }}} Similarly, the help for the "network import file" example from above is: {{{ help network import file network import file RootNetworkList=<ListSingleSelection [Create new network collection]> SourceColumnList=<ListSingleSelection [shared name]> TargetColumnList=<ListSingleSelection [shared name]> defaultInteraction=<String> delimiters=<ListMultipleSelection [,, ;, , \t]> delimitersForDataList=<ListSingleSelection [\|, \, /, ,]> filePath=<File> firstRowAsColumnNames=<boolean> indexColumnSourceInteraction=<int> indexColumnTargetInteraction=<int> indexColumnTypeInteraction=<int> startLoadRow=<int> }}} |
The Command Line Tool provides a simple command-line interface to Cytoscape using the Commands API. Any app that registers commands will be available through the Command Tool. The Command Tool provides two main functions: first, a Command Line Dialog is available from Tools → Command Line Dialog, that allows the user to type commands into Cytoscape and see the results in a "Reply Log".
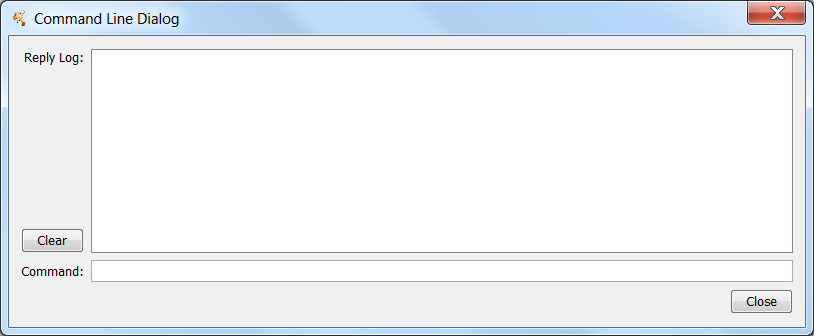
Second, and arguably more useful, it will read script files and execute them. Each line in the script file is a command that is sent to a app. Script files may be entered on the Cytoscape command line using the "-S" flag to Cytoscape, through the File → Run... menu item, or through Tools → Execute Command File.
Cytoscape commands consist of three parts: a command class, or namespace; a command within that namespace; and a series of arguments or options provided as a series of name=value pairs. For example, to import an XGMML format file from the Command Line Dialog or a command script, you would use:
network import file filePath="path-to-file"
where network is the namespace, import file is the command, and there is only one argument: filePath="path-to-file". If there were more arguments they would appear on the same line separated by spaces.
The Command Tool also uses the Command API to provide help. "help" by itself will list all of the command classes (or namespaces) and "help " followed by a namespace will list all of the commands supported by that namespace. Details of a specific command are available by typing "help " followed by the namespace and command (e.g. "help layout force-directed"). The Command Tool registers the "command" namespace and supports a single command: run, which takes a file argument. Here is the help for the command run command from the command namespace:
help command run
command run file=<File> Similarly, the help for the "network import file" example from above is:
help network import file
network import file RootNetworkList=<ListSingleSelection [Create new network collection]> SourceColumnList=<ListSingleSelection [shared name]> TargetColumnList=<ListSingleSelection [shared name]> defaultInteraction=<String> delimiters=<ListMultipleSelection [,, ;, , \t]> delimitersForDataList=<ListSingleSelection [\|, \, /, ,]> filePath=<File> firstRowAsColumnNames=<boolean> indexColumnSourceInteraction=<int> indexColumnTargetInteraction=<int> indexColumnTypeInteraction=<int> startLoadRow=<int>