|
← Revision 47 as of 2015-11-17 17:08:18
Size: 14003
Comment:
|
← Revision 48 as of 2016-04-26 22:15:22
Size: 61
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 1: | Line 1: |
|
## page was renamed from Cytoscape_3/UserManual/TableColumnData ## page was renamed from Cytoscape_User_Manual/TableColumnData ## page was renamed from Cytoscape_User_Manual/Attributes Interaction networks are useful as stand-alone models. However, they are most powerful for answering scientific questions when integrated with additional information. Cytoscape allows the user to add arbitrary node, edge and network information to Cytoscape as node/edge/network data columns. This could include, for example, annotation data on a gene or confidence values in a protein-protein interaction. These column data can then be visualized in a user-defined way by setting up a mapping from columns to network properties (colors, shapes, and so on). The section on '''[[Cytoscape_3/UserManual/Styles|Styles]]''' discusses this in greater detail. == Import Data Table Files == Cytoscape offers support for importing data from delimited text and MS Excel data tables. '''''Sample Data Table 1''''' ||Object Key||Alias||SGD ID|| ||AAC3||YBR085W|ANC3||S000000289|| ||AAT2||YLR027C|ASP5||S000004017|| ||BIK1||YCL029C|ARM5|PAC14||S000000534|| The data table file should contain a primary key column and at least one data column. The maximum number of data columns is unlimited. The '''Alias''' column is an optional feature, as is using the first row of data as column names. Alternatively, you can specify each column name from the '''File → Import → Table → File...''' user interface. === Basic Operation === {{attachment:ImportColumnsFromTable.png}} 1. Select '''File → Import → Table → File...'''. 1. Select a data file. The file can be either a text or Excel (.xls/.xlsx) file. 1. In the '''Target Table Data''' section, choose where to import the data to. You can choose an existing network collection, a specific network only, or you can choose to import the data to an '''Unassigned Table''' (described below). 1. Depending on what you choose in the '''Where to import Table Data''' drop-down, you will need to select a network collection or specific network. You will also need to select '''Importing Type''', that is whether the data is node, edge or network table columns. 1. If the table is not properly delimited in the preview panel, change the delimiter in the '''Advanced Options''' panel. The default delimiter is tab. This step is not necessary for Excel Workbooks. 1. By default, the first column is designated as the primary key. Change the key column if necessary. 1. Click '''OK''' to import. ==== Unassigned Table ==== As of Cytoscape 3.1. it is possible to import data tables without assigning them to existing networks, meaning the data doesn't have to correspond to any nodes/edges currently loaded. If a data table is imported as unassigned and a network is later imported that maps to the data in terms of nodes or edges, the data will link automatically. This is useful when loading a large dataset (for example expression data), defining a '''Style''' for visualizing the data on networks and later loading individual networks to view the data, for example from an online database. This feature allows the data to be automatically linked to any network that is applicable, without having to load the data for each network. == Legacy Cytoscape Attributes Format == In addition to tabular data, the simple attribute file format used in previous versions of Cytoscape is still supported. Node and edge data files are simply formatted: a node data file begins with the name of the column on the first line (note that it cannot contain spaces). Each following line contains the name of the node, followed by an equals sign and the data value. Numbers and text strings are the most common data types. All values for a given column must have the same type. For example: {{{ FunctionalCategory YAL001C = metabolism YAR002W = apoptosis YBL007C = ribosome }}} An edge data file has much the same structure, except that the name of the edge is the source node name, followed by the interaction type in parentheses, followed by the target node name. Directionality counts, so switching the source and target will refer to a different (or perhaps non-existent) edge. The following is an example edge data file: {{{ InteractionStrength YAL001C (pp) YBR043W = 0.82 YMR022W (pd) YDL112C = 0.441 YDL112C (pd) YMR022W = 0.9013 }}} Since Cytoscape treats edge data as directional, the second and third edge data values refer to two different edges (source and target are reversed, though the nodes involved are the same). Each data column is stored in a separate file. Node and edge data files use the same format, and have the suffix ".attrs". Node and edge data may be loaded via the '''File → Import → Table''' menu, just as data table files are. When expression data is loaded using an expression matrix, it is automatically loaded as node data unless explicitly specified otherwise. Node and edge data columns are attached to nodes and edges, and so are independent of networks. Data values for a given node or edge will be applied to all copies of that node or edge in all loaded network files, regardless of whether the data file or network file is imported first. === Detailed file format (Advanced users) === Every data file has one header line that gives the name of the data column, and optionally some additional meta-information about that data column. The format is as follows: {{{ columnName (class=JavaClassName) }}} The first field is always the column name: it cannot contain spaces. If present, the class field defines the name of the class of the data values. For example, java.lang.String or String for Strings, java.lang.Double or Double for floating point values, java.lang.Integer or Integer for integer values, etc. If the value is actually a list of values, the class should be the type of the objects in the list. If no class is specified in the header line, Cytoscape will attempt to guess the type from the first value. If the first value contains numbers in a floating point format, Cytoscape will assume java.lang.Double; if the first value contains only numbers with no decimal point, Cytoscape will assume java.lang.Integer; otherwise Cytoscape will assume java.lang.String. Note that the first value can lead Cytoscape astray: for example, {{{ floatingPointDataColumn firstName = 1 secondName = 2.5 }}} In this case, the first value will make Cytoscape think the values should be integers, when in fact they should be floating point numbers. It's safest to explicitly specify the value type to prevent confusion. A better format would be: {{{ floatingPointDataColumn (class=Double) firstName = 1 secondName = 2.5 }}} or {{{ floatingPointDataColumn firstName = 1.0 secondName = 2.5 }}} Every line past the first line identifies the name of an object (a node in a node data file or an edge in a edge data file) along with the String representation of the data value. The delimiter is always an equals sign; whitespace (spaces and/or tabs) before and after the equals sign is ignored. This means that your names and values can contain whitespace, but object names cannot contain an equals sign and no guarantees are made concerning leading or trailing whitespace. Object names must be the Node ID or Edge ID as seen in the left-most column of the Table Panel if the data column is to map to anything. These names must be reproduced exactly, including case, or they will not match. Edge names are all of the form: {{{ sourceName (edgeType) targetName }}} Specifically, that is ||sourceName space openParen edgeType closeParen space targetName|| Note that tabs are not allowed in edge names. Tabs can be used to separate the edge name from the "=" delimiter, but not within the edge name itself. Also note that this format is different from the specification of interactions in the SIF file format. To be explicit: a SIF entry for the previous interaction would look like {{{ sourceName edgeType targetName }}} or ||sourceName whiteSpace edgeType whiteSpace targetName|| To specify lists of values, use the following syntax: {{{ listDataColumnName (class=java.lang.String) firstObjectName = (firstValue::secondValue::thirdValue) secondObjectName = (onlyOneValue) }}} This example shows a data column whose value is defined as a list of text strings. The first object has three strings, and thus three elements in its list, while the second object has a list with only one element. In the case of a list every data value uses list syntax (i.e. parentheses), and each element is of the same class. Again, the class will be inferred if it is not specified in the header line. Lists are not supported by Styles and so can’t be mapped to network properties. === Newline Feature === Sometimes it is desirable to for data values to include linebreaks, such as node labels that extend over two lines. You can accomplish by inserting {{{\n}}} into the data value. For example: {{{ newlineDataColumn YJL157C = This is a long\nline for a label. }}} {{{#!wiki comment === Advanced Options === Is this still relevant? ==== Annotation File to Table Mapping ==== Formerly, Cytoscape only supported mapping between node/edge IDs and the primary keys in data files. With the introduction of Cytoscape 2.4, this limitation has been removed, and now both IDs and data columns with primitive data types (string, boolean, floating point, and integer) can be selected as the Key Attribute using the drop-down list provided. Complex attributes such as lists are not supported. {{attachment:attribute_table_import_keyattr.png}} ==== Aliases ==== Cytoscape uses a simple mechanism to manage aliases of objects. Both nodes and edges can have aliases. If an attribute is loaded as an alias, it is treated as a special attribute called "alias". This will be used when mapping attributes. If the primary key and key attribute for an object do not match, Cytoscape will search for a match between aliases and the key attribute. To define an alias column in the attribute table, just click on the checkboxes to the left of the column name while importing. {{attachment:attribute_table_import_alias.png}} }}} === Table Panel === {{attachment:TablePanel_withData.png}} When Cytoscape is started, the '''Table Panel''' appears in the bottom right of the main Cytoscape window. This browser can be hidden and restored using the F5 key or the '''View → Show/Hide Table Panel''' menu option. Like other Panels, the browser can be undocked by pressing the little icon in the top right corner. To swap between displaying node, edge, and network Data Tables use the tabs on the bottom of the Table Panel. By default, the Table Panel displays columns for all nodes and edges in the selected network. To display columns for only selected nodes/edges, click the '''Change Table Mode''' button {{attachment:TableMode.png}} at the top left. To change the columns that are displayed, click the '''Show Column''' {{attachment:ShowColumns.png}} button and choose the columns that are to be displayed (select various columns by clicking on them, and then click elsewhere on the screen to close the column list). Most column values can be edited by double-clicking the cell (only the ID cannot be edited). Newline characters can be inserted into String columns either by pressing '''Enter''' or by typing "\n". Once finished editing, click outside of the editing cell in the Table Panel or press '''Shift-Enter''' to save your edits. Pressing '''Esc''' while editing will undo any changes. Rows in the panel can be sorted alphabetically by specific column by clicking on a column heading. A new column can be created using the '''Create New column''' {{attachment:NewColumn.png}} button, and must be one of four types – integer, string, real number (floating point), or boolean. Columns can be deleted using the '''Delete Columns...''' {{attachment:DeleteColumns.png}} button. '''NOTE: Deleting columns removes them from Cytoscape, not just the Table Panel!''' To remove columns from the panel without deleting them, simply unselect the column using the '''Select Columns''' {{attachment:ShowColumns.png}} button. {{{#!wiki comment ==== Column Batch Editor ==== {{attachment:attribute_editor26.png}} The Attribute Browser has an '''Attribute Batch Editor'''. This enables you to set and modify attribute values for selected nodes or edges of a specified attribute at once. For example, if you want to create a new attribute called ''Modules'' and set module names for each group of selected nodes, you can use ''Set'' command from this editor. }}} == Import Data Table from Public Databases == It is also possible to import node data columns from public databases via web services, for example from BioMart. === Basic Operation === 1. Load a network, for example galFiltered.sif. 1. Select '''File → Import → Table → Public Databases...'''. 1. You will first be asked to select from a set of web services. For this example, we will choose '''ENSEMBL GENES 73 (SANGER UK)'''. {{attachment:select_services.png}} 1. In the '''Import Data Table from Web Services''' dialog, select a '''Data Source'''. Since galFiltered.sif is a yeast network, select '''ENSEMBL GENES - SACCHAROMYCES CEREVISIAE'''. 1. For '''Key Column in Cytoscape''', select ''shared name'' for '''Column''' and ''Ensembl Gene ID'' for '''Data Type'''. The type of identifier selected under '''Data Type''' must match what is used in the selected '''Column''' in the network. {{attachment:table_webservice.png}} 1. Select the data columns you want to import. 1. Select '''Import'''. When import is complete, you can see the newly imported data columns in the Table Panel. {{attachment:table_webservice_final.png}} |
#DEPRECATED <<Include(Cytoscape_3/UserManual/Deprecated)>> |
This is a legacy document
This page has been deprecated and is no longer updated. The current version of the Cytoscape manual can be found at http://manual.cytoscape.org/
Import Data Table Files
Cytoscape offers support for importing data from delimited text and MS Excel data tables.
Sample Data Table 1
Object Key |
Alias |
SGD ID |
AAC3 |
YBR085W|ANC3 |
S000000289 |
AAT2 |
YLR027C|ASP5 |
S000004017 |
BIK1 |
YCL029C|ARM5|PAC14 |
S000000534 |
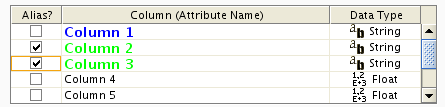
The data table file should contain a primary key column and at least one data column. The maximum number of data columns is unlimited. The Alias column is an optional feature, as is using the first row of data as column names. Alternatively, you can specify each column name from the File → Import → Table → File... user interface.
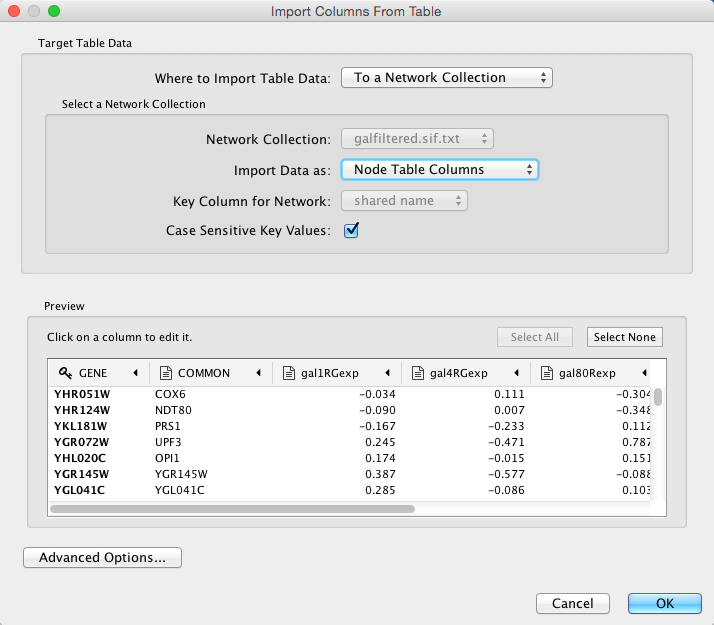
Basic Operation
Select File → Import → Table → File....
- Select a data file. The file can be either a text or Excel (.xls/.xlsx) file.
In the Target Table Data section, choose where to import the data to. You can choose an existing network collection, a specific network only, or you can choose to import the data to an Unassigned Table (described below).
Depending on what you choose in the Where to import Table Data drop-down, you will need to select a network collection or specific network. You will also need to select Importing Type, that is whether the data is node, edge or network table columns.
If the table is not properly delimited in the preview panel, change the delimiter in the Advanced Options panel. The default delimiter is tab. This step is not necessary for Excel Workbooks.
- By default, the first column is designated as the primary key. Change the key column if necessary.
Click OK to import.
Unassigned Table
As of Cytoscape 3.1. it is possible to import data tables without assigning them to existing networks, meaning the data doesn't have to correspond to any nodes/edges currently loaded. If a data table is imported as unassigned and a network is later imported that maps to the data in terms of nodes or edges, the data will link automatically. This is useful when loading a large dataset (for example expression data), defining a Style for visualizing the data on networks and later loading individual networks to view the data, for example from an online database. This feature allows the data to be automatically linked to any network that is applicable, without having to load the data for each network.
Legacy Cytoscape Attributes Format
In addition to tabular data, the simple attribute file format used in previous versions of Cytoscape is still supported. Node and edge data files are simply formatted: a node data file begins with the name of the column on the first line (note that it cannot contain spaces). Each following line contains the name of the node, followed by an equals sign and the data value. Numbers and text strings are the most common data types. All values for a given column must have the same type. For example:
FunctionalCategory YAL001C = metabolism YAR002W = apoptosis YBL007C = ribosome
An edge data file has much the same structure, except that the name of the edge is the source node name, followed by the interaction type in parentheses, followed by the target node name. Directionality counts, so switching the source and target will refer to a different (or perhaps non-existent) edge. The following is an example edge data file:
InteractionStrength YAL001C (pp) YBR043W = 0.82 YMR022W (pd) YDL112C = 0.441 YDL112C (pd) YMR022W = 0.9013
Since Cytoscape treats edge data as directional, the second and third edge data values refer to two different edges (source and target are reversed, though the nodes involved are the same).
Each data column is stored in a separate file. Node and edge data files use the same format, and have the suffix ".attrs".
Node and edge data may be loaded via the File → Import → Table menu, just as data table files are.
When expression data is loaded using an expression matrix, it is automatically loaded as node data unless explicitly specified otherwise.
Node and edge data columns are attached to nodes and edges, and so are independent of networks. Data values for a given node or edge will be applied to all copies of that node or edge in all loaded network files, regardless of whether the data file or network file is imported first.
Detailed file format (Advanced users)
Every data file has one header line that gives the name of the data column, and optionally some additional meta-information about that data column. The format is as follows:
columnName (class=JavaClassName)
The first field is always the column name: it cannot contain spaces. If present, the class field defines the name of the class of the data values. For example, java.lang.String or String for Strings, java.lang.Double or Double for floating point values, java.lang.Integer or Integer for integer values, etc. If the value is actually a list of values, the class should be the type of the objects in the list. If no class is specified in the header line, Cytoscape will attempt to guess the type from the first value. If the first value contains numbers in a floating point format, Cytoscape will assume java.lang.Double; if the first value contains only numbers with no decimal point, Cytoscape will assume java.lang.Integer; otherwise Cytoscape will assume java.lang.String. Note that the first value can lead Cytoscape astray: for example,
floatingPointDataColumn firstName = 1 secondName = 2.5
In this case, the first value will make Cytoscape think the values should be integers, when in fact they should be floating point numbers. It's safest to explicitly specify the value type to prevent confusion. A better format would be:
floatingPointDataColumn (class=Double) firstName = 1 secondName = 2.5
or
floatingPointDataColumn firstName = 1.0 secondName = 2.5
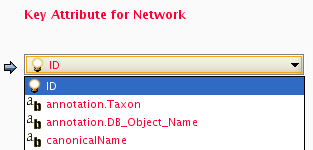
Every line past the first line identifies the name of an object (a node in a node data file or an edge in a edge data file) along with the String representation of the data value. The delimiter is always an equals sign; whitespace (spaces and/or tabs) before and after the equals sign is ignored. This means that your names and values can contain whitespace, but object names cannot contain an equals sign and no guarantees are made concerning leading or trailing whitespace. Object names must be the Node ID or Edge ID as seen in the left-most column of the Table Panel if the data column is to map to anything. These names must be reproduced exactly, including case, or they will not match.
Edge names are all of the form:
sourceName (edgeType) targetName
Specifically, that is
sourceName space openParen edgeType closeParen space targetName |
Note that tabs are not allowed in edge names. Tabs can be used to separate the edge name from the "=" delimiter, but not within the edge name itself. Also note that this format is different from the specification of interactions in the SIF file format. To be explicit: a SIF entry for the previous interaction would look like
sourceName edgeType targetName
or
sourceName whiteSpace edgeType whiteSpace targetName |
To specify lists of values, use the following syntax:
listDataColumnName (class=java.lang.String) firstObjectName = (firstValue::secondValue::thirdValue) secondObjectName = (onlyOneValue)
This example shows a data column whose value is defined as a list of text strings. The first object has three strings, and thus three elements in its list, while the second object has a list with only one element. In the case of a list every data value uses list syntax (i.e. parentheses), and each element is of the same class. Again, the class will be inferred if it is not specified in the header line. Lists are not supported by Styles and so can’t be mapped to network properties.
Newline Feature
Sometimes it is desirable to for data values to include linebreaks, such as node labels that extend over two lines. You can accomplish by inserting \n into the data value. For example:
newlineDataColumn YJL157C = This is a long\nline for a label.
Table Panel
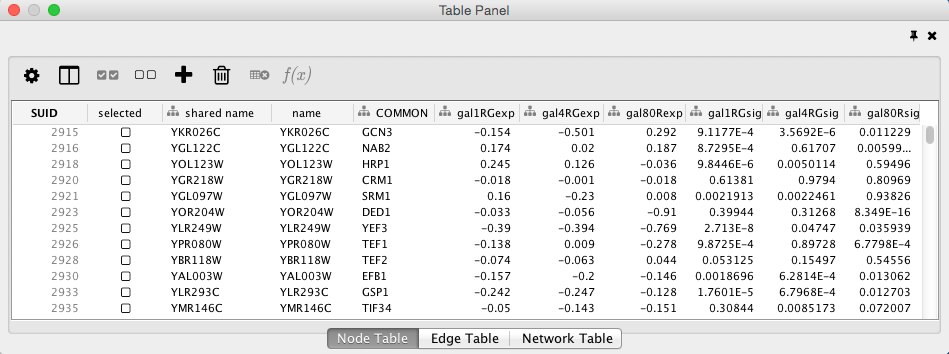
When Cytoscape is started, the Table Panel appears in the bottom right of the main Cytoscape window. This browser can be hidden and restored using the F5 key or the View → Show/Hide Table Panel menu option. Like other Panels, the browser can be undocked by pressing the little icon in the top right corner.
To swap between displaying node, edge, and network Data Tables use the tabs on the bottom of the Table Panel. By default, the Table Panel displays columns for all nodes and edges in the selected network. To display columns for only selected nodes/edges, click the Change Table Mode button  at the top left. To change the columns that are displayed, click the Show Column
at the top left. To change the columns that are displayed, click the Show Column  button and choose the columns that are to be displayed (select various columns by clicking on them, and then click elsewhere on the screen to close the column list).
button and choose the columns that are to be displayed (select various columns by clicking on them, and then click elsewhere on the screen to close the column list).
Most column values can be edited by double-clicking the cell (only the ID cannot be edited). Newline characters can be inserted into String columns either by pressing Enter or by typing "\n". Once finished editing, click outside of the editing cell in the Table Panel or press Shift-Enter to save your edits. Pressing Esc while editing will undo any changes.
Rows in the panel can be sorted alphabetically by specific column by clicking on a column heading. A new column can be created using the Create New column  button, and must be one of four types – integer, string, real number (floating point), or boolean. Columns can be deleted using the Delete Columns...
button, and must be one of four types – integer, string, real number (floating point), or boolean. Columns can be deleted using the Delete Columns...  button. NOTE: Deleting columns removes them from Cytoscape, not just the Table Panel! To remove columns from the panel without deleting them, simply unselect the column using the Select Columns
button. NOTE: Deleting columns removes them from Cytoscape, not just the Table Panel! To remove columns from the panel without deleting them, simply unselect the column using the Select Columns  button.
button.
Import Data Table from Public Databases
It is also possible to import node data columns from public databases via web services, for example from BioMart.
Basic Operation
- Load a network, for example galFiltered.sif.
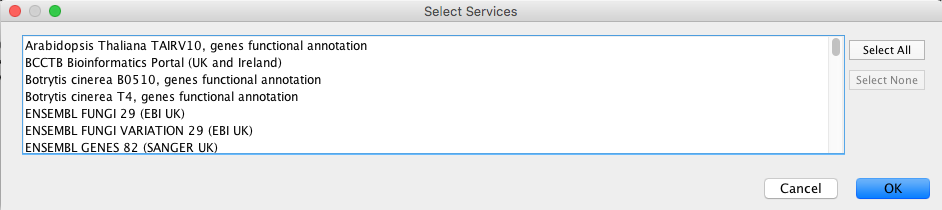
Select File → Import → Table → Public Databases....
You will first be asked to select from a set of web services. For this example, we will choose ENSEMBL GENES 73 (SANGER UK).
In the Import Data Table from Web Services dialog, select a Data Source. Since galFiltered.sif is a yeast network, select ENSEMBL GENES - SACCHAROMYCES CEREVISIAE.
For Key Column in Cytoscape, select shared name for Column and Ensembl Gene ID for Data Type.
The type of identifier selected under Data Type must match what is used in the selected Column in the network.
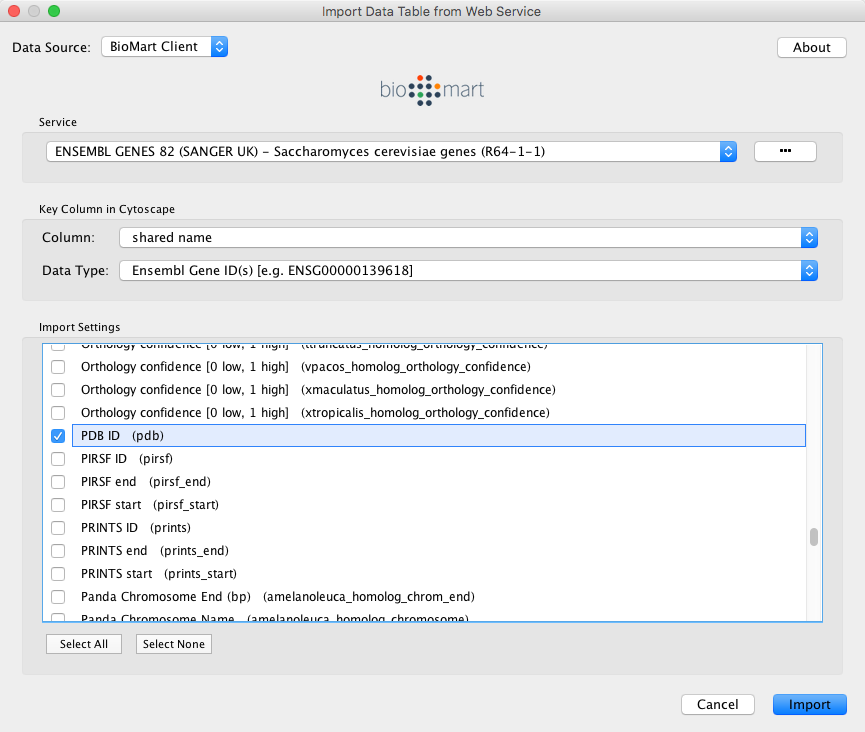
- Select the data columns you want to import.
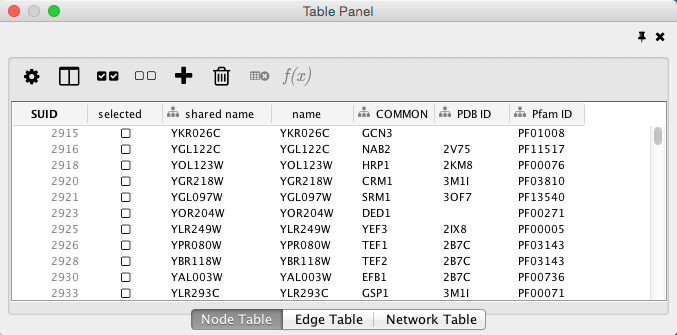
Select Import.
When import is complete, you can see the newly imported data columns in the Table Panel.
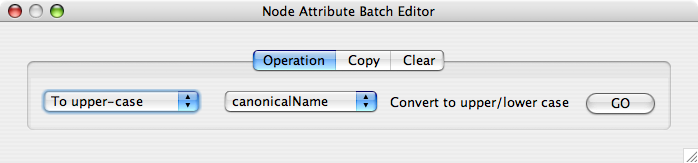 The Attribute Browser has an Attribute Batch Editor. This enables you to set and modify attribute values for selected nodes or edges of a specified attribute at once. For example, if you want to create a new attribute called Modules and set module names for each group of selected nodes, you can use Set command from this editor.
The Attribute Browser has an Attribute Batch Editor. This enables you to set and modify attribute values for selected nodes or edges of a specified attribute at once. For example, if you want to create a new attribute called Modules and set module names for each group of selected nodes, you can use Set command from this editor.