|
← Revision 15 as of 2015-01-21 01:25:01
Size: 6482
Comment: fixed relative links
|
← Revision 16 as of 2017-05-29 18:21:32
Size: 6384
Comment: added app-dev link
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 46: | Line 46: |
| * The [[https://groups.google.com/forum/#!forum/cytoscape-discuss | Discussion Mailing List]] is for users, app developers and core developers. | * The [[https://groups.google.com/forum/#!forum/cytoscape-app-dev | App Developer Mailing List]] is for asking questions about app development. |
| Line 48: | Line 48: |
| * The [[https://groups.google.com/forum/#!forum/cytoscape-helpdesk | Help Desk]] is where users can ask questions on usage and app development. | * The [[https://groups.google.com/forum/#!forum/cytoscape-helpdesk | Help Desk]] is where users can ask questions on usage. |
| Line 51: | Line 51: |
| * [[https://www.pinterest.com/cytoscape/ | Cytoscape Figures on Pinterest]] |
Introduction to Cytoscape 3.0 App Development
Overview
This step will give you an introduction to what Cytoscape is, how it is used, and an overview of technologies used.
What is Cytoscape?
Cytoscape is an open source software platform for visualizing molecular interaction networks and biological pathways and integrating these networks with annotations, gene expression profiles and other state data. Although Cytoscape was originally designed for biological research, now it is a general platform for complex network analysis and visualization. Cytoscape core distribution provides a basic set of features for data integration, analysis, and visualization. Additional features are available as Apps. Apps are available for network and molecular profiling analyses, new layouts, additional file format support, scripting, and connection with databases. They may be developed by anyone using the Cytoscape open API based on Java™ technology and App community development is encouraged. Most of the Apps are freely available from http://apps.cytoscape.org/.
What is Cytoscape used for?
Cytoscape is a general network analysis tool, and can therefor be used to analyze network data from many areas of research:
Biology
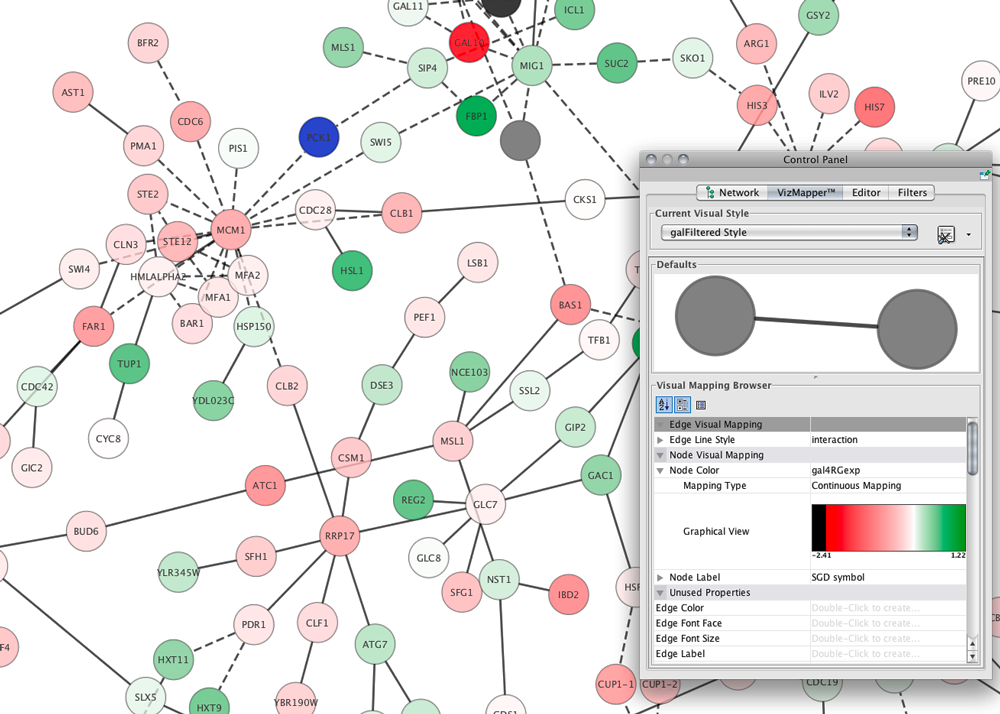
Cytoscape supports many use cases in molecular and systems biology, genomics, and proteomics:
- Load molecular and genetic interaction data sets in many standards formats
- Project and integrate global datasets and functional annotations
- Establish powerful visual mappings across these data
- Perform advanced analysis and modeling using Cytoscape Apps
Visualize and analyze human-curated pathway datasets such as WikiPathways, Reactome, and KEGG
Social Science
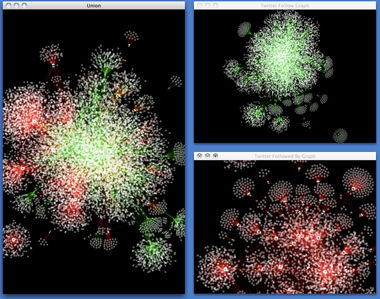
Cytoscape is used by social scientists to:
- Visualize and analyze large social networks of interpersonal relationships
- Assemble social networks from tables and forms
- Gather social interactions from the web by variety of web service APIs with scripting languages and save it in standard data file formats. Cytoscape supports most of the standard file formats
- Calculate network statistics using Apps
- Use with other tools, such as R with sna/ igraph package or NetworkX, for more advanced analysis
General Complex Network Analysis
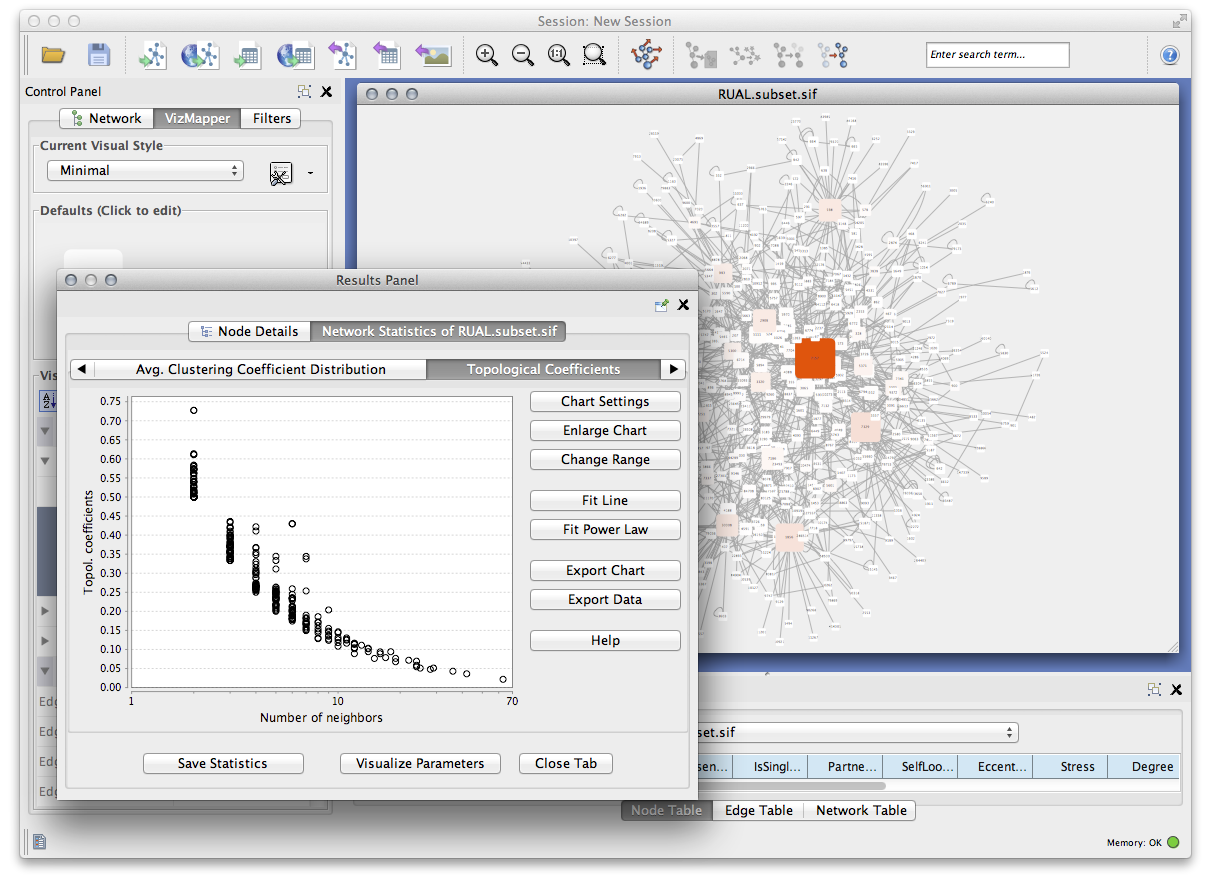
Cytoscape is domain-independent and therefore is a powerful tool for complex network analysis in general:
Calculate statistics for networks by Apps such as NetworkAnalyzer or CentiScaPe
- Find shortest path
- Find clusters by various kinds of algorithms
- Use with other tools for more advanced analysis
Perform advanced network analysis in popular tools, including igraph, Pajek, or GraphViz and import it to Cytoscape as standard file formats like GraphML
The Cytoscape Community
Cytoscape has an active and diverse community of developers and users. Communication happens mostly via mailing lists and social media:
The App Developer Mailing List is for asking questions about app development.
The Announcement Mailing List is used for announcements from the core development team to the rest of the community.
The Help Desk is where users can ask questions on usage.
The Cytoscape Publications Tumblr highlights published research using Cytoscape and published plugins.
Cytoscape Architecture and Technologies
Cytoscape 3.0 has a clearly defined, simplified API. API jars are strictly separate from the implementation jars. We believe that we’ve learned our lessons with 2.x and have greatly improved the usability of the API to benefit our active developer community. The API is versioned using the Semantic Versioning standard. This means that the API won't change throughout 3.x, so an app designed to work with an early version of 3.x will be guaranteed to work up to version 4.0 of Cytoscape. Our commitment to supporting app developers is stated in an explicit backwards compatibility contract found in each class in the public API so that both core developers and app writers will understand how a class might change.
Cytoscape 3.0 strives for a modular architecture where the application consists of a set of jar files where each jar contributes a well defined subset of functionality to the application. The design is oriented around the OSGi service model (i.e. micro service) with API and implementation separated into different jar files. We’ve introduced the use of OSGi as a way to create and enforce a modular, sustainable code base. We’ve also introduced Maven as a build system to help manage the many jar files created. Read more about these
The Relationship Between Cytoscape Core and Apps
Cytoscape core includes a set of basic features for import, export, layout and analysis of networks and associated data. Specialized and advanced features are available Apps distributed through the Cytoscape App Store. To date, over 200 Apps are available. If you've developed Cytoscape plugins/apps before, then you will notice quite a few changes in developing for Cytoscape 3.0. These changes cover both core and plugin/app architectures. To highlight the differences and added benefits of developing on the Cytoscape 3.0 platform, we're now calling them apps. So, consider porting your 2.x plugins to 3.0 apps. If you are new to Cytoscape development, please ignore plugins and focus on the app developer documentation.