|
Size: 2477
Comment:
|
← Revision 13 as of 2009-02-12 01:03:14
Size: 2518
Comment: converted to 1.6 markup
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 1: | Line 1: |
| [[TableOfContents([])]] | <<TableOfContents([])>> |
| Line 48: | Line 48: |
| 1. Apply layouts for each networks | |
| Line 50: | Line 51: |
| attatchment:merge1.png | {{attachment:merge1.png}} |
<<TableOfContents: execution failed [Argument "maxdepth" must be an integer value, not "[]"] (see also the log)>>
Introduction
This is a tutorial how to merge multiple networks from multiple data sources by Advanced Network Merge plugin.
Examples
Setup
You need to install following plugins to try this tutorial.
Required
- Advanced Network Merge Plugin
PathwayCommons web service client
IntAct web service client
- NCBI Entrez Gene web service client
- GPML Plugin
- Biomart Client plugin
Optional
- Scripting Engine Manager
Scinario 1: Merge existing network file and network imported from web service
This is the most common usecase for this plugin. Supporse users have their own data sets (networks) and want to integrate them with other network data sets available from public databases. This plugin can easily handle this problem.
In this example, we are going to use human network data sets (rual.sif) and expand this network from public databases.
Data Source
- User network (sif, XGMML, etc.)
- Known interaction data sets from public database
- NCBI Entrez Gene - interaction datasets are taken from HPRD, BioGRID, and BIND
- Annotations from public resources
- Biomart
- NCBI Entrez Gene
Procedure
- Start Cytoscape. Assume you have already installed all required plugins.
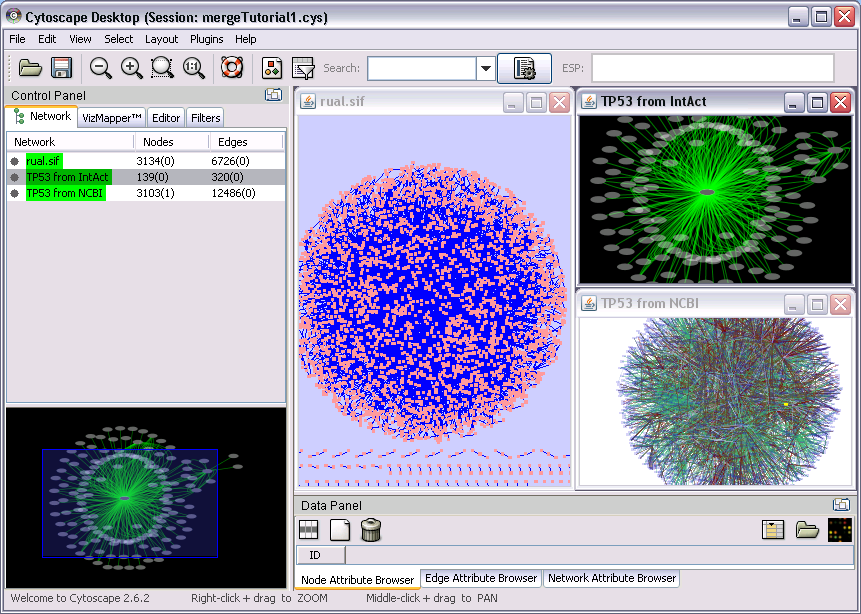
Import user network. In this example, use RUAL.sif in the example. Import > Network and select URL. From the plull-down menu, select Rual et al. (rual.sif) dataset. Then press Import
Select Import > Network from Web Services...
For Data Source, select IntAct Web Service Client
Type TP53 AND human in the Query field, then press Search
When you asked to enter network name, type TP53 from IntAct
Select Import > Network from Web Services...
For Data Source, select NCBI Entrez Eutilities Web Service Client
Type TP53 AND human then press search.
Name the network (TP53 from Entrez)
- Apply layouts for each networks
Select View > Arrange Network Windows > Tiled. At this point, your workspace looks like the following:
(Under construction)
Search
Scinario 2: Merge known pathways onto human interactome data set
Data Source